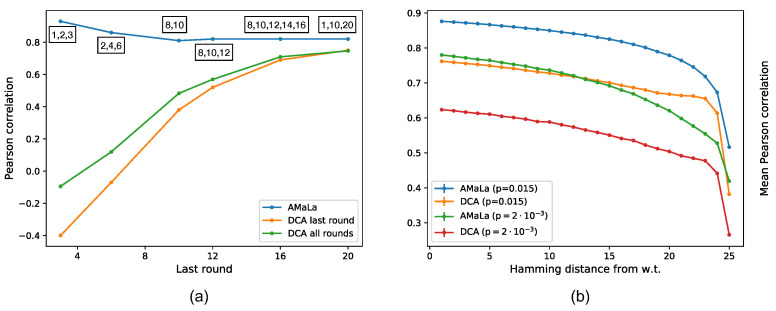
Figure 5.
Inferred signal dependence on the number of rounds and hamming distance from wild-type. Left panel (a): Pearson correlation when the number of rounds are changing. Comparison between PlmDCA on the last round (or all rounds) and AMaLa. The sequenced rounds are: (1, 2, 3); (2, 4, 6); (8, 10); (8, 10, 12); (8, 10, 12, 14, 16); (1, 10, 20). PlmDCA significantly depends on the number of performed rounds, not significantly inferring the fitness landscape up to round . On the contrary, AMaLa provides predicted energy functions highly correlated with the fitness even for a low number of performed selection rounds. Right panel (b): Degradation of the mean Pearson correlation between inferred and true energies as a function of the Hamming distance from the wild-type sequence. Two different simulations are considered: high () and low () mutation probability. Changing such parameters varies the broadness of the library screening during an experiment, resulting in probing a more local or more broad region of the sequence space. AMaLa predictions are systematically better than PlmDCA, while the latter displays a slower decrease in correlation augmenting the distance from wild-type.