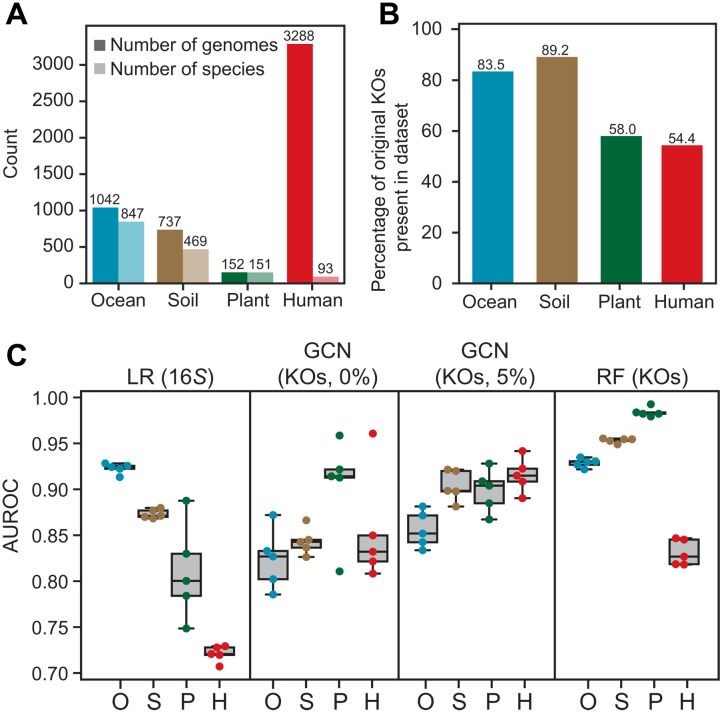
Fig. 5. Predictions of HGT are accurate for small ecology–specific datasets.
(A) The number of genomes and species in each of four datasets: ocean surface sampling, soil microbial communities, rhizomes from three plant species, and human gut microbiomes from 11 individuals. (B) The number of KOs overlapping with the HGT model (shown in Fig. 1B) used for predictions. (C) Model performance, defined by AUROC, for HGT predictions within the ocean, soil, plant and human gut microbiome datasets, using either the LR model based on 16S rRNA distances, the GCN model using gene functions (KOs) with either none or 5% of the network exposed, or the RF model using gene functions (KOs).