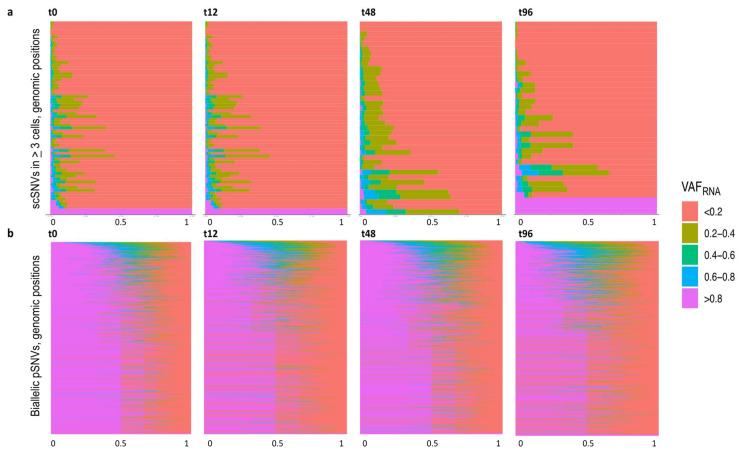
Figure 4.
(a). ScVAFRNA estimated at positions covered by a minimum of 3 sequencing reads (minR = 3) for sceSNVs called in 3 and more cells per dataset (y-axis). The majority of the positions have a VAFRNA up to 0.2. Note that the plot is inclusive for all the cells with minR = 3 in the corresponding position, including those covered with reference reads only. The percentage of cells with a corresponding VAFRNA is displayed on the x-axis. (b). ScVAFRNA estimated at those positions covered by a minimum of 3 sequencing reads for biallelic pSNVs (y-axis). For most of the pSNVs, the VAFRNA distribution is centered around 0.5, which is expected for germline heterozygous SNVs not subjected to monoallelic expression.