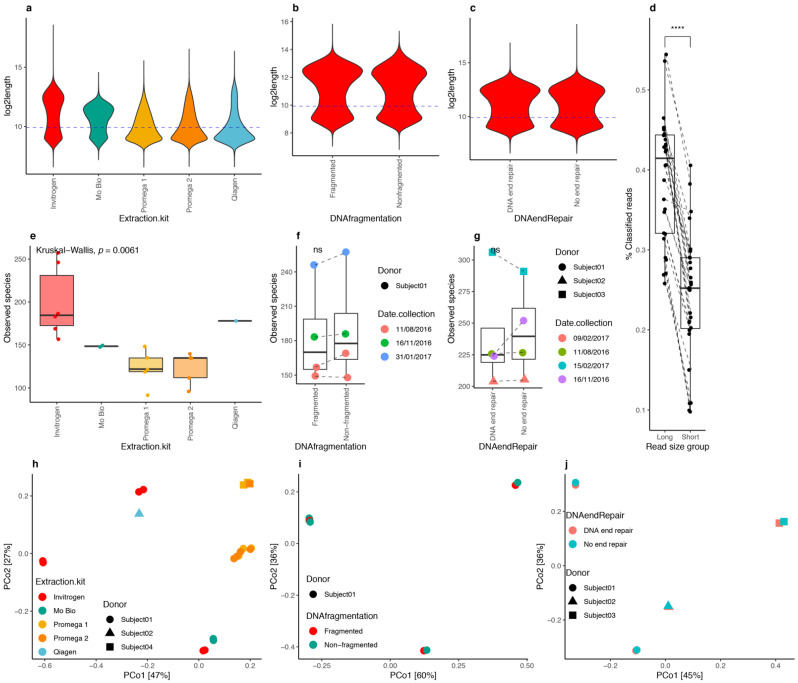
Figure 4.
DNA extraction kits, fragmentation, and end-repair impact on human stool metagenomic composition from ONT sequencing data. Read length distributions of ONT reads across different DNA extraction kits ((a) n = 29) and between DNA fragmentation ((b) n = 6 paired samples fragmented/nonfragmented) and DNA end repair ((c) n = 6 paired samples end vs. no end repair) steps for Invitrogen samples. Blue dashed lines correspond to the median value of log2-transformed read lengths used to stratify reads as long or short. (d) Differences between the fraction of classified reads by the Centrifuge approach between long and short reads for 29 samples in panel (a). (e) Differences in microbial diversity (observed species) between extraction kits (n = 29). (f) Differences in microbial diversity (observed species) by DNA fragmentation (n = 4 paired samples). (g) Differences in microbial diversity (observed species) by DNA end-repair step (n = 4 paired samples). (h) PCoA ordination of 29 samples in panel (a) colored by extraction kit. (i) PCoA ordination of 8 samples in panel (f). (j) PCoA of 8 samples of panel (g) colored by DNA end-repair step. ns (panel (f,g)) = nonsignificant differences in paired Wilcoxon rank-sum tests. **** = p-value < 0.0001 in paired Wilcoxon rank-sum test.