To the Editor:
Bone is a common site of metastasis in breast cancer. Due to discordance between biomarker expression in primary and metastatic tumors, biopsies of bone metastases are performed prior to treatment to reassess estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) to guide treatment (1). Decalcifying agents used to enable sectioning of bone biopsies affect the accuracy and feasibility of ER, PR, and HER2 determinations using immunohistochemistry (IHC) and fluorescence in situ hybridization by negatively impacting antigenicity and DNA quality (2). A more robust technique for evaluating expression of key receptors could help to ensure optimal treatment. We assessed the feasibility of extracting protein from non-decalcified bone biopsies and quantifying ER, PR, and HER2 via proteotypic peptides using peptide immunoenrichment-multiple reaction monitoring-mass spectrometry (immuno-MRM-MS). We previously described an immuno-MRM-MS assay for quantifying ER and HER2 from cell lysates and tissues (3).
Under an IRB-approved protocol, we obtained core needle biopsies of bone metastasis from 16 patients with breast cancer (Harborview Medical Center, Seattle, Washington). Samples from separate needle pulls were analyzed by immuno-MRM-MS and pathology, which could cause variability in intra-patient samples, yet would still allow an assessment of the feasibility of the immuno-MRM-MS approach.
All samples were analyzed in a blinded fashion by immuno-MRM-MS. Wet tissue weight ranged from 3–195 mg. Samples were cryofractured without decalcification, and protein was extracted using a standard proteomic urea lysis buffer. Protein yield ranged from 63–5,554 μg (mean percent yield was 2.2%, range 0.6–6.4%). Protein lysates were digested using trypsin (9 samples for which >300 μg protein/sample was available were aliquoted into triplicates), and a mixture of monoclonal antibodies targeting proteotypic peptides from ER, PR, and HER2 was covalently-linked to magnetic beads and used in multiplex to enrich the endogenous and spiked-in stable isotope labeled standard (SIS) peptides prior to immuno-MRM-MS. Specificity during MRM was confirmed by: i) co-elution of SIS and endogenous peptides, ii) monitoring 5 fragment ions/peptide, and iii) consistent relative peak areas for light and SIS fragment ions (Fig. 1A). For each peptide, 3–4 interference-free fragment ions were used for quantitative analysis. Expression levels of endogenous peptides were reported as light-to-heavy peak area ratios, normalized by protein input. Limits of detection (LODs) for ER, PR, and HER2 were 0.001, 0.01, and 0.007 light-to-heavy peak area ratios per mg, respectively, determined based on three times the signal-to-noise ratio in negative control samples (100 μg of breast cancer cell line T-47D; ER+, PR+, HER2+), spiked with SIS peptides but without antibodies.
Figure 1. Quantification of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) in bone biopsies by immuno-MRM-MS compared with immunohistochemistry (IHC).
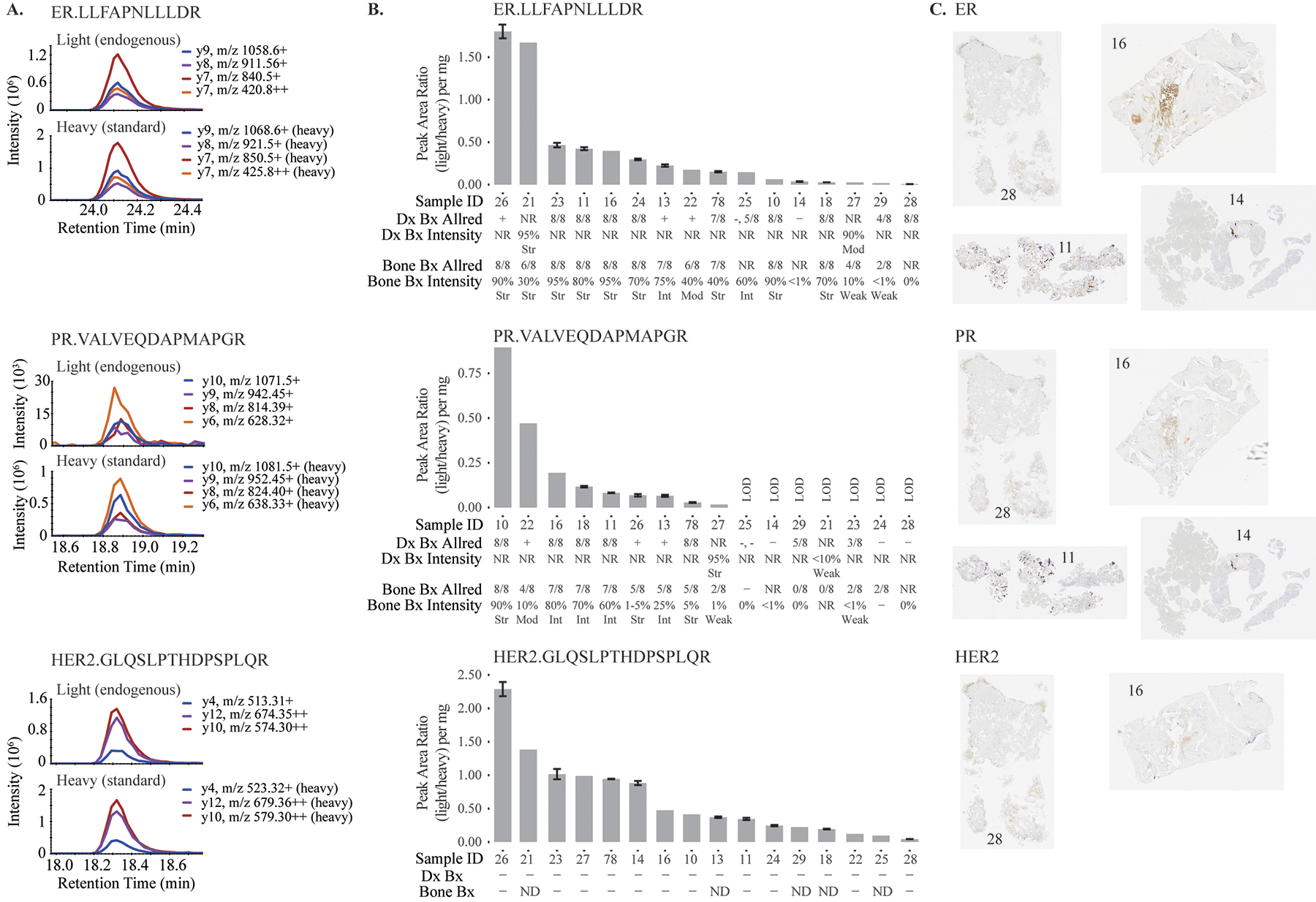
A. Chromatograms illustrating peptide specificity. B. Immuno-MRM-MS measurements and respective pathology biomarker status results of the primary diagnostic biopsies (Dx Bx) and the metastatic bone biopsies (Bone Bx). Error bars represent standard deviations for process triplicates. C. Representative IHCs. NR=not reported, Str=strong, Mod=moderate, Int=intermediate, ND=not determinable due to decalcification; LOD=not detected above noise limit.
Endogenous ER, PR, and HER2 peptides were detected above the LODs in 16, 9, and 16 samples, respectively. The median %CVs for measurements above LOD (for process triplicates) were 5.1% (1.3–9.8%) for ER, 7.7% (1.8–10.1%) for PR, and 3.5% (0.5–7.6%) for HER2. The pathology reports and immuno-MRM-MS measurements showed substantial agreement, and it is noteworthy that HER2 could be quantified by immuno-MRM-MS in patients where IHC failed to make a determination (Fig. 1B). For ER, 11 IHC-positive or intermediate biopsies also had the 11 highest immuno-MRM-MS results, and three IHC-negative samples were among the five lowest samples by immuno-MRM-MS (Fig. 1B, 1C). Two samples were discordant, possibly due to the pathology samples’ originating from separate needle pulls than the immuno-MRM-MS samples, or subjective IHC interpretation for sample 27 (10% of tumor cells stained weakly, Allred 4/8). For PR, eight IHC-positive or intermediate samples also had the eight highest immuno-MRM-MS measurements, and eight IHC-negative samples correlated with the lowest immuno-MRM-MS measurements or no PR peptide detection. For HER2, although the clinical assays did not identify any positive samples, immuno-MRM-MS enabled quantification of HER2 in all 16 samples, consistent with reports that HER2 protein expression can be detected in samples found negative by IHC (4). Notably, for 5 of the 16 patients, the HER2 status could not be determined after decalcification when using conventional methods, but HER2 was successfully quantified by immuno-MRM-MS.
Further analytical characterization of the assay in bone matrix and a larger clinical validation study are needed to determine thresholds for clinical use, but these proof-of-principle data demonstrate the feasibility of quantifying biomarkers from non-decalcified bone biopsies using immuno-MRM-MS, providing a basis for further investigation of the clinical utility of the approach. Peptide immuno-MRM-MS is currently reflexively used in several clinical laboratories to quantify serum thyroglobulin in patients with autoantibodies that interfere with immunometric assays, demonstrating the feasibility of harmonizing such assays across laboratories (5). Of note, because MRM-based assays are multiplexable, expression levels of multiple druggable targets could be assessed alongside ER, PR, and HER2, potentially identifying patients who may respond to targeted therapies.
Author contributions / Acknowledgements:
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved. R. M. Schoenherr processed samples, acquired data, and wrote the manuscript; U. Voytovich processed samples and acquired data; J. R. Whiteaker helped with manuscript writing and data interpretation; V. K. Gadi enrolled patients and reviewed data; J. R. Gralow and A. G. Paulovich conceived and coordinated the study, and A. G. Paulovich contributed to manuscript writing.
We acknowledge enrolling physicians (William Gwin, Kalyan Banda, Jennifer Specht, Hannah M. Linden, Sasha Stanton, Larissa Korde, Rachel Yung), project/regulatory management contributors (Kim Dammann, Jenna Olyanich, Obsy Tadesse), sample and data retrieval contributors (Lena Rubinstein, Kathleen Hanson, Madelyn Peha, Delaney Niebusch, Melissa Sharkey), and sample processing and data analysis contributors (Lei Zhao, Jacob J. Kennedy, ChenWei Lin, Laura Kennedy).
Research Funding:
The research reported here was supported by the Seattle Cancer Consortium Breast SPORE (P50CA138293) and the Safeway Foundation, the National Cancer Institute (NCI) of the National Institutes of Health (NIH) Clinical Proteomics Tumor Analysis Consortium Initiative (nos. U24CA160034 and U01CA214114), the Innovative Molecular Analysis Technologies (IMAT) Program (no. R33CA173300), the Academic-Industrial Partnership (AIP) Program (no. R01CA235575), and the NCI Research Specialist Program (no. R50CA211499).
Abbreviations:
- IHC
immunohistochemistry
- immuno-MRM-MS
peptide immunoenrichment-multiple reaction monitoring-mass spectrometry
- SIS
stable isotope labeled standard peptides
- LOD
limit of detection
Footnotes
Authors’ Disclosures or Potential Conflict of Interest:
The authors declare no conflicts of interest.
References
- 1.Van Poznak C, Somerfield MR, Bast RC, Cristofanilli M, Goetz MP, Gonzalez-Angulo AM, et al. Use of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American society of clinical oncology clinical practice guideline. J Clin Oncol 2015;33:2695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schrijver WA, van der Groep P, Hoefnagel LD, Ter Hoeve ND, Peeters T, Moelans CB, van Diest PJ. Influence of decalcification procedures on immunohistochemistry and molecular pathology in breast cancer. Mod Pathol 2016;29:1460–70. [DOI] [PubMed] [Google Scholar]
- 3.Schoenherr RM, Whiteaker JR, Zhao L, Ivey RG, Trute M, Kennedy J, et al. Multiplexed quantification of estrogen receptor and Her2/Neu in tissue and cell lysates by peptide immunoaffinity enrichment mass spectrometry. Proteomics 2012;12:1253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim HK, Park KH, Kim Y, Park SE, Lee HS, Lim SW, et al. Discordance of the pam50 intrinsic subtypes compared with immunohistochemistry-based surrogate in breast cancer patients: Potential implication of genomic alterations of discordance. Cancer Res Treat 2019;51:737–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Netzel BC, Grant RP, Hoofnagle AN, Rockwood AL, Shuford CM, Grebe SK. First steps toward harmonization of LC-MS/MS thyroglobulin assays. Clin Chem 2016;62:297–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


