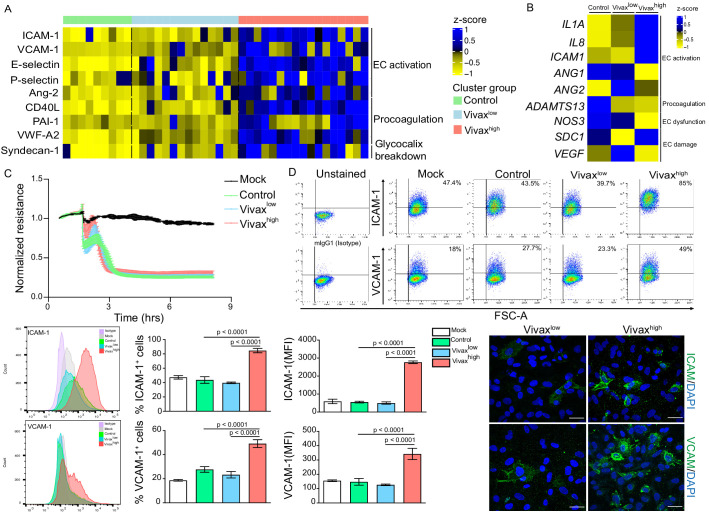
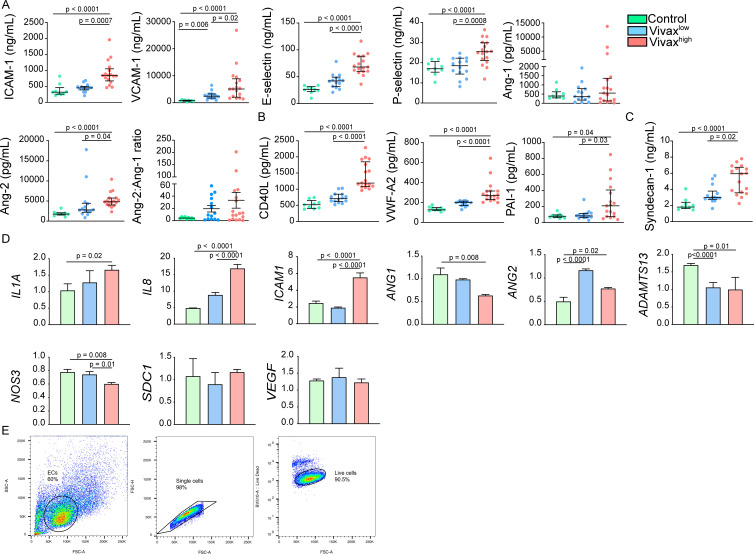
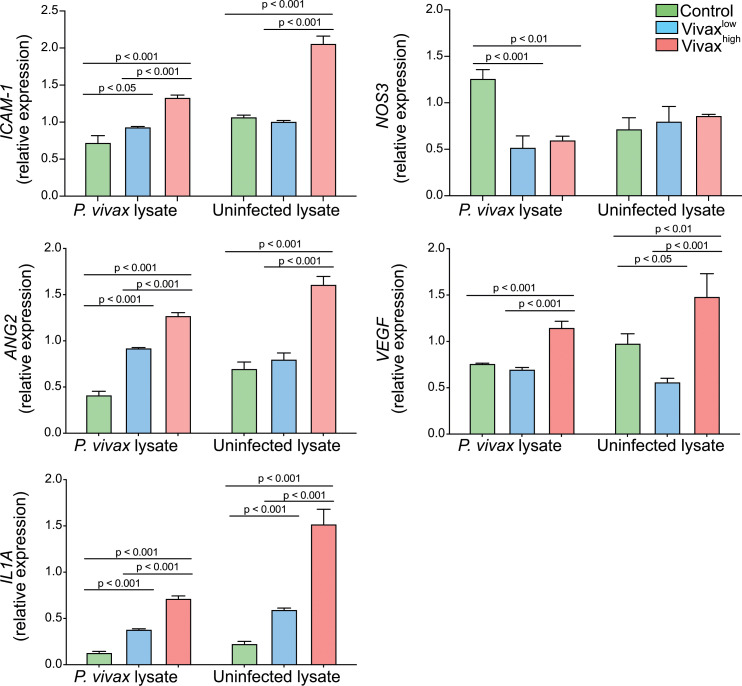
Figure 5. Elevated circulating markers of endothelial cell (EC) activation and damage in Vivaxhigh compared to Vivaxlow patients.
(A) Endothelial changes across clusters: Luminex. Heatmap represents z-scores obtained by centering values of Luminex data. Shown are markers of EC activation, procoagulation, and glycocalyx damage. Each column (each individual) in the heatmap is matched with colour-coded cluster assignment: Cluster Control – green bar; Cluster Vivaxlow – blue bar; and Cluster Vivaxhigh – red bar. (B) Endothelial changes across clusters: qRT-PCR. Transcriptional response of human umbilical vein endothelial cells (HUVECs) incubated for 6 hr with 30% v/v pooled plasma from different clusters. Heatmap reflects relative mRNA expression intensity (average scaled value) after results were normalized to GAPDH housekeeping gene expression and untreated condition (mock). Data shown represent the mean of three independent experiments. For each experiment, two technical replicates were performed for each condition. (C) Endothelial changes across clusters: impedance changes. Endothelial monolayer integrity was measured during 20% v/v of pooled plasma incubation. Each line represents the mean ± SD of normalized resistance of HUVECs measured by electric cell-substrate impedance sensing (ECIS) at 4000 Hz. Data shown are representative of three independent experiments. For each experiment, two technical replicates were performed for each condition. (D) Endothelial changes across clusters: imaging and flow cytometry. HUVECs were incubated for 18 hr with 30% v/v of pooled plasma of individuals in the different clusters or left untreated (mock). Percentage of cells expressing EC activation markers (adhesion molecules) ICAM and VCAM as well as quantification of protein expression was determined by flow cytometry and immunofluorescence analysis (scale bar = 33 μM). Isotype antibodies were used as control to define positive populations. Significance was calculated for comparisons between conditions at the corresponding time point . One-way analysis of variance statistical test with Tukey’s corrected multiple comparisons test was performed. p-Value is indicated above the graph when p<0.05. Data shown are representative mean ± SEM of three independent experiments.