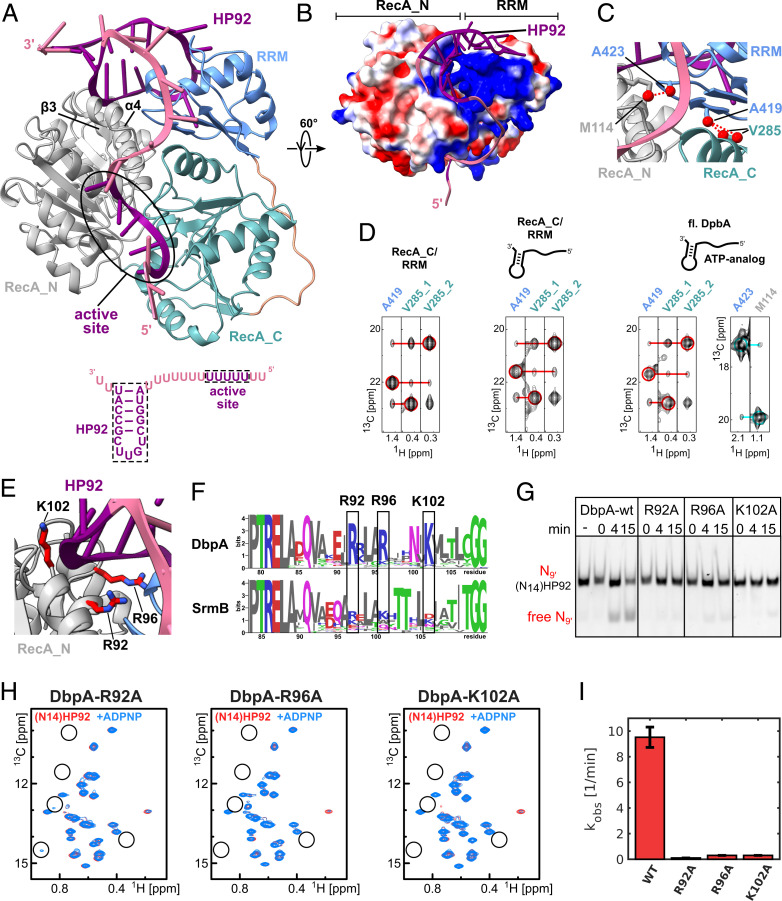
Fig. 4.
A model of the closed state of DbpA reveals stabilizing interactions between the RecA_N domain and HP92 that enable helicase activity. (A) The model of the closed state based on the solution structure of the RecA_C/RRM construct shows that HP92 and the RRM contact the RecA_N domain in the closed state. HP92 and 5 nt of the 5′ ss overhang bound to the ssRNA binding site of the helicase core were modeled based on previously published structures (3, 17). These regions are shown in purple, and the rest of the RNA is shown in pale violet. RRM, RecA_N and RecA_C domains are shown in blue, gray, and green, respectively. β-3 and α-4 of the RecA_N domain that interact with HP92 and the RRM are indicated. The RNA construct that is bound to the helicase in the model is shown on the bottom. (B) Surface representation the modeled DbpA colored according to the electrostatic surface potential (positive, blue; negative, red). The structure is rotated by 60° compared to A. The stem of HP92 packs into a positively charged groove formed between RRM and RecA_N domains. (C) Closeup of the interdomain interface of the closed state. Methyl groups that are predicted to show strong interdomain NOEs (dashed, red lines) are shown in red. The short distance between V285 and A419 (Right) is used to validate the orientation between the RecA_C and RRM domains. The short distance between A423 and M114 reports on the correct orientation between the RecA_N and RRM domains. (D) CCH-NOESY strips along the 13C-NOE dimension that show the interdomain NOEs for the RecA_C/RRM construct in the free form (Left), bound to (N14)HP92 RNA (Middle), or for full length DbpA in the closed state (Right). Interdomain NOEs between the RecA_C (V285) and RRM domains (A419) are indicated by red lines. The interdomain NOE between the RecA_N and RRM domains in the closed state (between A423 and M114) is indicated by a cyan line. Diagonal peaks are indicated by circles. (E) Closeup of the interaction between the RecA_N domain and HP92 in the closed state. The conserved Arg/Lys residues (R92/R96/K102) are shown in stick representation (red). (F) Sequence logos for DbpA (Top) and the unrelated DEAD-box helicase SrmB (Bottom). Residues R92, R96, and K102 are highly conserved across DbpA homologs, whereas the equivalent positions in SrmB are not conserved. The PTRELAXQ (Left) and GG motifs (Right) belong to the characteristic sequence motifs of DEAD-box helicases. (G) Mutation of the conserved Arg/Lys residues abolishes the helicase activity of DbpA. Unwinding of a fluorescently labeled 9mer RNA hybridized to (N14)HP92 RNA is followed by native PAGE after 0-, 4-, and 15-min incubation time. DbpA-wt shows robust unwinding after 15-min incubation time; no unwinding is observed after 15 min for the wt in the absence of ATP (−). Neither of the DbpA-R92A, -R96A, and -K102A mutants shows significant unwinding after 15 min. (H) Ile region of the methyl TROSY spectra of the DbpA-R92A, -R96A, and -K102A mutants bound to (N14)HP92 RNA (red) and after addition of ADPNP (blue). The positions of some of the characteristic signals of the closed state are indicated by circles. The mutations impair the formation of the closed conformation (R96A and K102A) or strongly reduce the population of the closed conformation (R92A) (compare to Fig. 2B). (I) Comparison of the ATPase activity of DbpA-wt with the -R92A, -R96A, and -K102A mutants. All measurements were performed in the presence of (N14)HP92 RNA. The mutations strongly reduce the ATPase activity of DbpA.