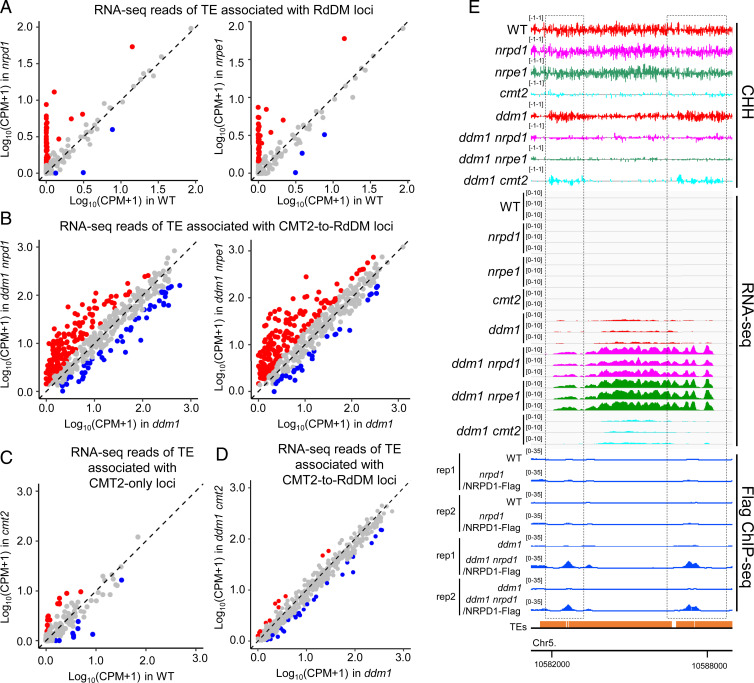
Fig. 3.
The CMT2-to-RdDM pathway suppresses the expression of TEs in ddm1. (A–D) Scatterplot showing RNA-seq reads of the indicated TEs in the indicated genotypes. CPMs were averaged from three biological replicates. Each dot represents a single TE. Red dots represent TEs up-regulated in the genotype in the y axis compared with genotype in the x axis (fold change >2, FDR <0.05), whereas blue dots represent TEs down-regulated (fold change >2, FDR <0.05). TEs are considered as associated with the indicated loci when their body or flanking 2-kb regions overlap with the indicated loci. RNA-seq of cmt2 and WT are from ref. 14. (E) CHH methylation levels, TE expression levels, and NRPD1 ChIP-seq signals at a representative region from CMT2-to-RdDM repressed TEs. Black dotted boxes indicate the CMT2-to-RdDM loci. The numbers in parentheses indicate y axis scales.