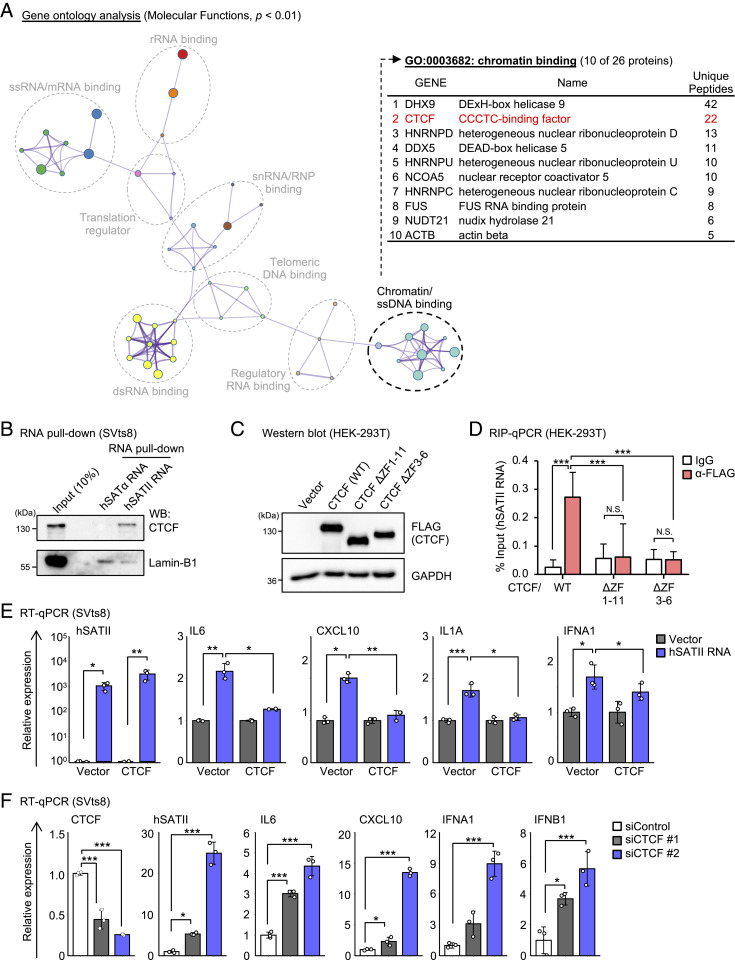
Fig. 2.
Pericentromeric hSATII RNA binds to CTCF. (A) GO analysis of 280 hSATII RNA–binding proteins (Left). Among these proteins, 26 genes were categorized as chromatin-binding (GO: 0003682), and the top 10 ranked genes and unique peptides are listed (Right). (B) RNA pull-down assay using SVts8 cell lysate followed by Western blotting confirmed hSATII RNA but not hSATα RNA bound to CTCF. (C) Western blot analysis of FLAG-tagged CTCF (WT: wild type) or CTCF ΔZF (deletion of ZF domain) in HEK-293T cells. (D) RIP followed by qPCR confirmed the binding of FLAG-tagged CTCF WT, but not CTCF ΔZF1-11 or ΔZF3-6, to hSATII RNA in HEK-293T cells. (E) RT-qPCR analysis of SASP-like inflammatory genes in hSATII RNA–overexpressed SVts8 cells with excess CTCF. The relative expression shows the value normalized from empty vector–expressed cells. (F) RT-qPCR analysis of SASP-like inflammatory genes in CTCF-depleted SVts8 cells. The relative expression shows the value normalized from small-interfering control (siControl)–treated cells. Each bar represents mean ± SD of three technical replicates repeated in two independent experiments (D, E, and F). *P < 0.05, **P < 0.01, ***P < 0.001, or N.S. (not significant) by one-way ANOVA, followed by the Tukey’s (D and E) or Dunnett’s (F) multiple comparisons post hoc test.