Significance
Class A penicillin-binding proteins (aPBPs) assemble the bacterial cell wall and are the targets of penicillin and related β-lactam antibiotics. In gram-negative bacteria, the aPBPs require outer membrane lipoproteins to function. However, little is known about how these proteins promote the activity of their cognate synthases in cells. Here, we show that one of these lipoproteins, called LpoA, has a much more pronounced effect on aPBP activity in cells than anticipated from biochemical studies. It not only modulates the cross-linking of cell wall polymers but is also required for the aPBP to make the polymers in the first place. Our findings therefore provide insights into the regulation of an important class of antibiotic targets in their native cellular context.
Keywords: peptidoglycan, penicillin, lipoprotein, cell wall, outer membrane
Abstract
A cell wall made of the heteropolymer peptidoglycan (PG) surrounds most bacterial cells. This essential surface layer is required to prevent lysis from internal osmotic pressure. The class A penicillin-binding proteins (aPBPs) play key roles in building the PG network. These bifunctional enzymes possess both PG glycosyltransferase (PGT) and transpeptidase (TP) activity to polymerize the wall glycans and cross-link them, respectively. In Escherichia coli and other gram-negative bacteria, aPBP function is dependent on outer membrane lipoproteins. The lipoprotein LpoA activates PBP1a and LpoB promotes PBP1b activity. In a purified system, the major effect of LpoA on PBP1a is TP stimulation. However, the relevance of this activation to the cellular function of LpoA has remained unclear. To better understand why PBP1a requires LpoA for its activity in cells, we identified variants of PBP1a from E. coli and Pseudomonas aeruginosa that function in the absence of the lipoprotein. The changes resulting in LpoA bypass map to the PGT domain and the linker region between the two catalytic domains. Purification of the E. coli variants showed that they are hyperactivated for PGT but not TP activity. Furthermore, in vivo analysis found that LpoA is necessary for the glycan synthesis activity of PBP1a in cells. Thus, our results reveal that LpoA exerts a much greater control over the cellular activity of PBP1a than previously appreciated. It not only modulates PG cross-linking but is also required for its cognate synthase to make PG glycans in the first place.
The peptidoglycan (PG) cell wall is an essential structure surrounding most bacterial cells. It provides them with their characteristic shape and protects their cytoplasmic membrane from osmotic lysis. PG is composed of glycan strands with a repeating unit of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) connected by a β-1 to -4 linkage (1). A peptide stem is attached to the MurNAc sugars of the polymer and is used to form cross-links between adjacent glycans, generating an interconnected matrix that encases the cell (1). Many of our best antibiotics, including penicillin and related β-lactam drugs, target the assembly of this structure (2). Thus, in addition to addressing a fundamental biological problem, understanding the molecular mechanisms underlying PG biogenesis and its regulation also promises to aid the development of novel antibacterial therapies.
PG is built from a lipid-linked precursor called lipid II, which consists of a disaccharide-peptide monomer unit attached to an undecaprenol lipid via a pyrophosphate linkage (1). Once the lipid II precursor has been synthesized and transported to the outer face of the cytoplasmic membrane, it is polymerized and cross-linked into the PG matrix by enzymes with PG glycosyltransferase (PGTase) and transpeptidase (TPase) activity, respectively (1). There are two main types of PG synthases. The most well studied of the two are the class A penicillin-binding proteins (aPBPs), which have a large extracytoplasmic domain with both PGTase and TPase subdomains (3). The second type of synthase was discovered more recently and is formed by a SEDS (shape, elongation, division, sporulation) family PGTase in complex with a monofunctional, class B PBP (bPBP) with TPase activity (4–7). Although it remains a matter of debate how these two types of synthases work together to build the PG layer, the SEDS-bPBP complexes are known to be the essential enzymes of the cell elongation and division machineries in most bacteria (4, 5, 7). The aPBPs, on the other hand, are thought to play critical roles in maintaining the integrity of the PG matrix (5, 8–10).
In Escherichia coli, the aPBP-type synthases PBP1a and PBP1b form a synthetic lethal pair (11, 12). Mutants inactivated for either enzyme alone are viable, whereas the simultaneous loss of both of their activities results in rapid cell lysis. In E. coli and other gram-negative bacteria, aPBP function requires interaction with outer membrane lipoproteins (13–17). LpoA is the activator for PBP1a, and this pairing is widely conserved among the gamma-proteobacteria (13, 14). In contrast, the LpoB activator of PBP1b has a more limited phylogenetic distribution with the unrelated LpoP protein substituting for it in Pseudomonas aeruginosa and many other members of the gamma-proteobacteria (13, 14, 17). In purified systems, the lipoproteins activate both the PGT and TP activities of their cognate synthases (13, 14, 18–20). However, LpoB and LpoP have a much more profound effect on the PG polymerase activity of their cognate PBP1b proteins than on cross-linking (18–20). In contrast, the major effect of LpoA on PBP1a is the stimulation of TPase activity (19). Unlike LpoB, the effect of LpoA on the PGTase activity of its cognate synthase is completely abolished by the inhibition of TP activity with β-lactams (19). Based on these biochemical results, the primary function of LpoB and LpoP is thought to be the activation of the PGTase activity of PBP1b, and the major function of LpoA is thought to be the activation of the TPase activity of PBP1a (1).
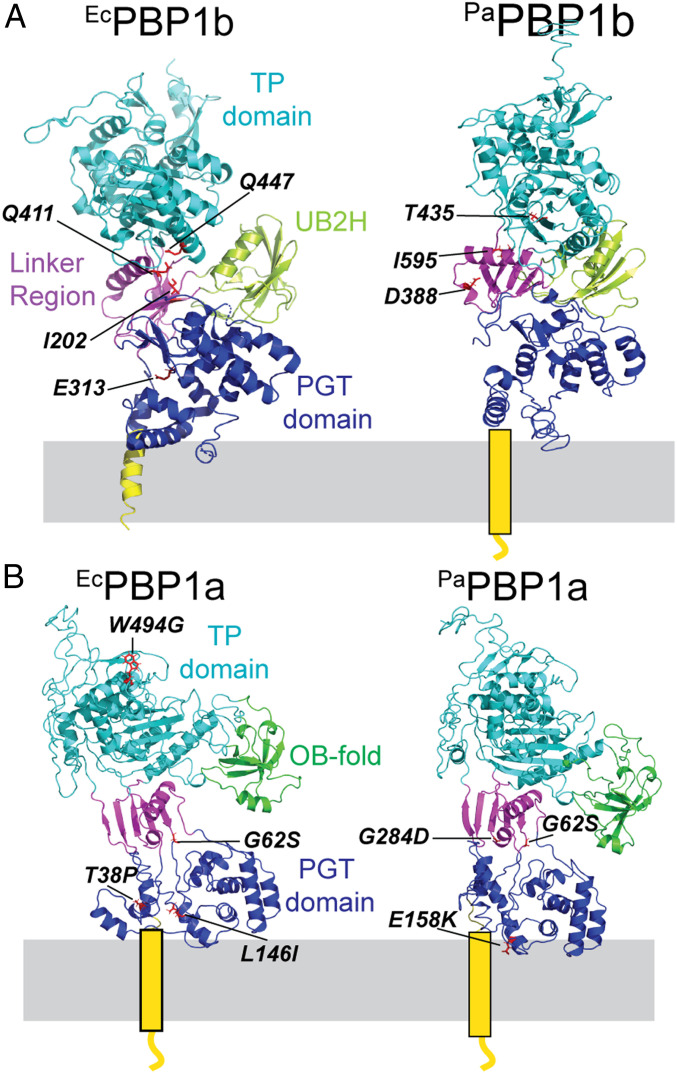
To investigate the physiological significance of the activation of PBP1b by LpoB in cells, we previously identified E. coli PBP1b variants that bypassed the requirement for LpoB activation in vivo (21). The amino acid substitutions resulting in LpoB bypass activity mapped either to the PGTase domain or clustered in the interdomain linker region near the LpoB-binding domain (UB2H domain) of the PBP1b structure (Fig. 1A). Purified PBP1b variants with these changes had enhanced PGTase activity. Notably, variants of P. aeruginosa PBP1b that bypass the requirement for LpoP activation map to a similar region in the modeled structure of this enzyme (Fig. 1A) (17). The observation that PBP1b variants with elevated PGTase activity bypass the lipoprotein requirement for cellular function provides strong support for the in vivo activity of LpoB and LpoP being the activation of PG polymerization by PBP1b.
Fig. 1.
PBP structures and locations of activator bypass substitutions. Shown is the structure of E. coli PBP1b (PDB ID code 3VMA) (35) (A) and model structures of P. aeruginosa PBP1b (A), E. coli PBP1a (B), and P. aeruginosa PBP1a (B) made using i-Tasser. The PBP1a structures were based on the structure from Acinetobacter baumannii (PDB ID code 3UDF) (37). The different domains of the PBPs are color-coded. The locations of the previously identified LpoB/LpoP bypass mutations in PBP1b (17) (A) are indicated on the structures as are the LpoA bypass substitutions in PBP1a identified in this report (B).
Although the effect of LpoA on PBP1a activity has been extensively studied in vitro, the role of LpoA in the activation of its cognate PBP in cells has remained unclear. There are many reasons to believe that LpoA may have a distinct function from that of LpoB and act on its target PBP in a different way. The two lipoproteins are structurally and evolutionarily divergent (14, 18, 22–24). They also bind different accessory domains on their cognate aPBP and stimulate different PG synthetic activities in vitro (Fig. 1B) (14, 19). Therefore, to learn more about the cellular role of LpoA, we selected for variants of PBP1a that can function in the absence of the activator in both E. coli and P. aeruginosa. Similar to the PBP1b bypass variants (21), PBP1a bypass variants have amino acid substitutions that cluster in the interdomain linker region of PBP1a and hyperactivate its PGTase activity. Thus, this domain appears to play a conserved role in modulating the synthesis of glycan strands by different types of gram-negative aPBP enzymes. Using an in vivo assay for PGTase activity (5, 25, 26), we further showed that PBP1a requires LpoA to promote PG polymerization in cells. Thus, our results reveal that LpoA exerts a much greater control over the cellular activity of PBP1a than previously appreciated. It not only modulates PG cross-linking but is also required to activate PG polymerization by its cognate synthase. Furthermore, based on the similar locations of the lipoprotein bypass substitutions, our findings suggest that LpoA and LpoB regulate the cellular activity of their cognate aPBPs in a fundamentally similar way despite differences in their structure and in vitro behavior.
Results
Identification of E. coli PBP1a Variants That No Longer Require LpoA for Activity.
E. coli strain MM20 has its native ponB gene encoding PBP1b engineered to be under control of the arabinose promoter (Para::ponB). It also has a deletion of lpoA and therefore cannot survive in the absence of arabinose because both aPBPs are inactivated in this growth condition: PBP1b by depletion and PBP1a due to its missing activator. To identify PBP1a variants that can bypass the requirement for LpoA (designated as PBP1a* variants), we transformed MM20 [ΔlpoA Para::ponB] with a plasmid library harboring a PCR-mutagenized allele of the PBP1a-encoding gene ponA under control of the isopropyl β-d-thiogalactopyranoside (IPTG)-inducible lactose promoter (Plac::ponA). Plasmids encoding PBP1a* variants were expected to promote the survival of MM20 in the absence of arabinose due to the ability of the altered synthases to supply vital aPBP activity to the PBP1b-depleted cells that lack LpoA. The MM20 transformants were therefore plated on LB agar without arabinose to deplete PBP1b, but with IPTG to induce PBP1a production. Plasmids were then isolated from the surviving colonies and transformed back into the parental background to confirm that the LpoA bypass phenotype was plasmid-linked. The ponA gene from such plasmids was then sequenced to identify the mutation conferring bypass activity.
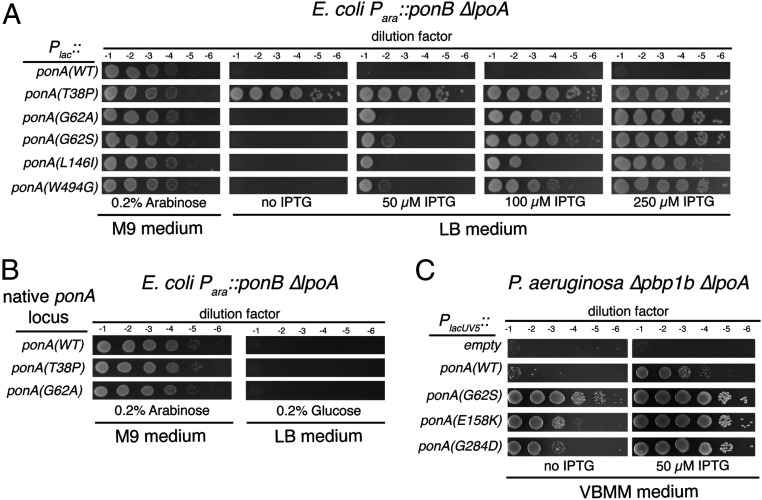
Five ponA* alleles with single base-pair changes were identified that we chose to characterize further. These mutants encoded PBP1a(T38P), PBP1a(G62S), PBP1a(G62A), PBP1a(L146I), and PBP1a(W494G). Given that LpoA has principally been found to stimulate the TP activity of PBP1a in vitro, it was notable that only one of the substitutions in the PBP1a* candidates mapped to the TP domain with the remainder clustering in the PGT domain and linker region between the two catalytic domains (Fig. 1B). To confirm their LpoA bypass activity, the genes encoding these variants were transferred to a new parental vector backbone and reintroduced into strain MM20 [ΔlpoA Para::ponB]. All of the alleles restored the growth of the strain upon PBP1b depletion (Fig. 2A). However, all PBP1a* variants except for PBP1a(T38P) required elevated inducer concentrations to achieve LpoA bypass (Fig. 2A). Additionally, allelic replacement of the two strongest alleles, ponA(T38P) and ponA(G62S), at the native ponA locus in strain MM20 [ΔlpoA Para::ponB] failed to promote growth in the absence of arabinose (Fig. 2B). Thus, the identified PBP1a* variants indeed bypass the requirement for LpoA but they require overproduction from a plasmid to serve as the sole functioning aPBP in the absence of the activator.
Fig. 2.
PBP1a* variants bypass the need for LpoA for function in vivo. (A). Overnight cultures of MM20 [∆lpoA Para::ponB] containing the plasmids pJLB16 [Plac::ponA], pJLB20 [Plac::ponA(G62A)], pJLB21 [Plac::ponA(G62S)], pJLB22 [Plac::ponA(L146I)], pJLB25 [Plac::ponA(W494G)], or pJLB29 [Plac::ponA(T38P)] were grown in M9-arabinose-Cam medium at 30 °C. Cells were then pelleted, washed once in M9 salts, and resuspended in M9 salts to an OD600 of 1.0. The cell suspensions were then serially diluted, and 5 μL of each dilution was spotted onto M9-arabinose-Cam agar or LB-Cam agar containing IPTG, as indicated. Plates were incubated at 30 °C overnight prior to imaging. Note that we use M9 arabinose as our permissive condition because we find that the Para::ponB construct is expressed better in this medium than in LB. (B) Overnight cultures of MM20 [∆lpoA Para::ponB], JLB128 [∆lpoA ponA(G62S) Para::ponB], and JLB127 [∆lpoA ponA(T38P) Para::ponB] were grown and serially diluted as in A, except the medium lacked Cam. The dilutions were spotted onto M9-arabinose or LB-glucose agar as indicated. Plates were incubated at 30 °C overnight prior to imaging. (C). Overnight cultures of PA686 cells [∆ponB ∆lpoA] harboring the plasmids pPSV38 [PlacUV5::empty], pNG91 [PlacUV5::ponA], pNG94 [PlacUV5::ponA(E158K)], pNG95 [PlacUV5::ponA(G62S)], or pNG96 [PlacUV5::ponA(G284D)] were grown in LB no salt with Gent at 30 °C. Cells were then pelleted, washed twice in an equal volume of VBMM media, and resuspended in VBMM at an OD600 of 1.0. The cell suspensions were then diluted and plated as above on VBMM agar plates containing the indicated concentration of IPTG. Plates were incubated at 37 °C overnight prior to imaging.
Identification of PBP1a* Variants in P. aeruginosa.
To determine whether similar substitutions in PBP1a would also bypass its LpoA requirement in other bacteria, we sought to identify P. aeruginosa PBP1a* variants. As in E. coli, PBP1a and PBP1b form a synthetic lethal pair in P. aeruginosa (17). However, in the case of P. aeruginosa, a mutant deleted for both ponB and lpoA is conditionally viable. It can grow on LB medium but not on Vogel–Bonner minimal medium (VBMM) (17). We were therefore able to select for mutants encoding PBP1a* variants by plating the ∆lpoA ∆ponB strain on VBMM. Spontaneous suppressors of the VBMM plating defect were isolated and subjected to whole-genome sequencing. Three mutants were identified with single base-pair changes in the ponA gene. They encoded the PBP1a variants PBP1a(G62S), PBP1a(E158K), and PBP1a(G284D), which like the LpoA bypass substitutions identified in E. coli, map to the PGT and interdomain linker region (Fig. 1B). Strikingly, one of the variants, PBP1a(G62S), had a bypass substitution identical to that identified in the E. coli protein. To confirm the LpoA bypass activity of the identified variants, their corresponding genes were cloned into multicopy plasmids under the control of an IPTG-inducible promoter. They were then introduced into the ∆lpoA ∆ponB strain and tested for their ability to promote growth on VBMM medium. In the absence of inducer, the empty vector and the plasmid encoding ponA(WT) both failed to suppress the VBMM growth phenotype of the ∆lpoA ∆ponB strain (Fig. 2C). In contrast, the growth defect in the absence of inducer was partially suppressed by the plasmids encoding the PBP1a variants, with PBP1a(G284D) displaying the weakest suppression activity (Fig. 2C). In the presence of a low concentration of inducer, all three mutant alleles robustly suppressed the VBMM growth defect, whereas the defect was only partially suppressed by ponA(WT) (Fig. 2C). We therefore conclude that amino acid substitutions in the PGT and interdomain region of PBP1a confer LpoA bypass activity to synthases from both E. coli and P. aeruginosa. Thus, the mechanism of PBP1a activity regulation in gram-negative bacteria is likely to be conserved.
E. coli PBP1a* Variants Have Elevated PGT and TP Activity In Vitro.
To understand how the E. coli PBP1a* variants bypass the requirement for LpoA in cells, we investigated the effect of the identified amino acid substitutions on PBP1a activity in vitro. The four variants—PBP1a(T38P), PBP1a(G62S), PBP1a(L146I), and PBP1a(W494G)—were overexpressed as His6-SUMO–tagged fusion proteins, purified by Ni-NTA affinity chromatography, and the tag was removed with SUMO protease. Unfortunately, the PBP1a(W494G) variant did not stably accumulate in cells when it was overexpressed, preventing the characterization of its biochemical activity.
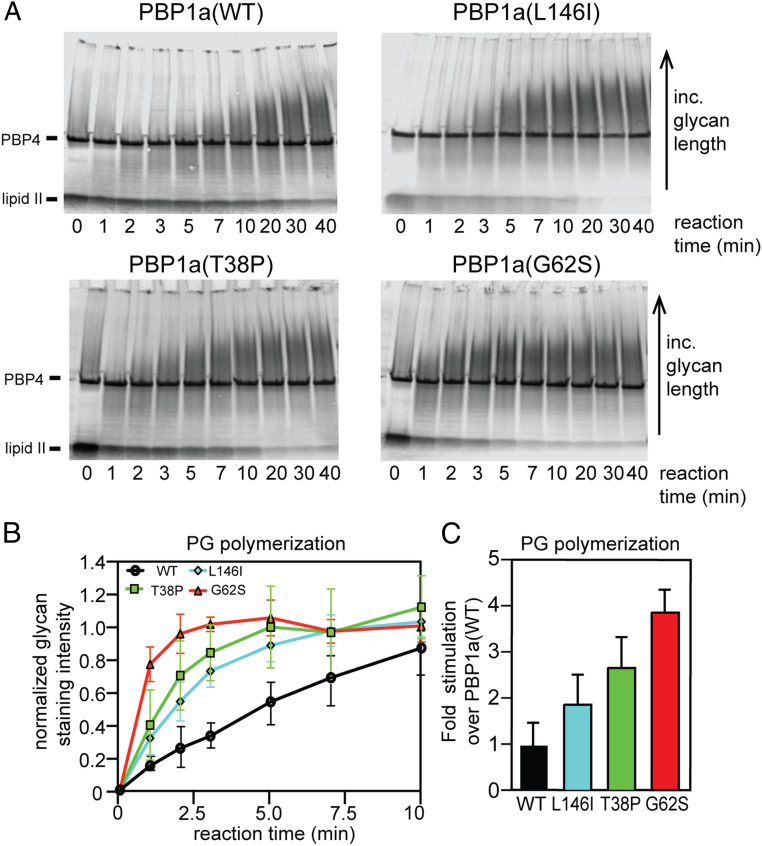
We first chose to assess the PGT activity of the remaining variants relative to the wild-type protein. The formation of glycan strands was monitored over time after incubation of the PBP1a derivatives with lipid II substrate purified from Enterococcus faecalis followed by biotinylation and detection of the polymeric products using recently developed methods (27–30). We used the E. faecalis substrate because it cannot be cross-linked by E. coli PBPs due to L-Lys being present at position 3 of the peptide instead of meso-diaminopimelic acid (mDAP), which is the native residue found in E. coli lipid II. This property allowed the size of PG polymerization products to be monitored by gel electrophoresis without complications from cross-linking. Additionally, polymers derived from the E. faecalis substrate are labeled more effectively with biotinylated-d-lysine by Staphylococcus aureus PBP4 (28), allowing better product detection than if the native E. coli substrate was used. All three of the PBP1a* variants displayed elevated PGT activity relative to the wild-type enzyme. Although PBP1a(T38P) appeared to be the most active variant in vivo, the PBP1a(G62S) variant was the most active PG polymerase in vitro (Fig. 3). The reason for the differential behavior of the PBP1a(T38P) variant between the in vivo and in vitro assays is not known, but it may be due to poor conformational stability of the protein when it is removed from the membrane or purified away from other potential protein partners in the cell.
Fig. 3.
E. coli PBP1a* variants have increased glycosyltransferase activity. (A) Purified PBP1a and its variants (5 µM) were incubated with purified E. faecalis lipid II (40 µM) for the indicated times. Reactions were stopped by incubation at 95 °C for 2 min. Products were incubated with S. aureus PBP4 (1 µM) and biotin-d-lysine (800 µM) to label the PG glycans produced. Labeled samples were then separated on an SDS/PAGE gel, transferred to a PVDF membrane, and visualized with IRDye 800CW Streptavidin. (B) The intensity of the glycan staining was quantified for each lane of each time series in A and plotted. Error bars represent the SD based on five replicates [PBP1a and PBP1a(G62S)], two replicates [PBP1a(T38P)], or three replicates [PBP1a(L146I)]. (C) Shown are the activities of each PBP1a variant relative to PBP1a(WT) at the 2-min time point. Activities were calculated by setting the activity for one of the PBP1a(WT) replicates to 1.0 and then calculating the activity of the other samples relative to it. Error bars represent the SD of the mean.
Purified LpoA reacted with the biotinylated-d-lysine used for labeling the PG glycan products, resulting in a strongly labeled band in the gel-based PGT assay. The significance of this labeling is not clear, and the high background signal it generated unfortunately prevented us from being able to test the effect of the lipoprotein on the PGT activity of PBP1a using this assay. Thus, we could not compare the relative effect of the PBP1a* substitutions on PGT activity to the stimulation induced when LpoA is added to PBP1a(WT). However, the activation of the PGTase activity observed for the PBP1a(G62S) variant (4-fold) was higher than the stimulation of wild-type PBP1a by LpoA observed previously using a radiolabeled lipid II substrate (1.5-fold) (19). Thus, the biochemical results indicate that changes in PBP1a resulting in the LpoA bypass phenotype result in elevated in vitro PG polymerase activity with the level of stimulation higher than that induced by coincubation of PBP1a with LpoA.
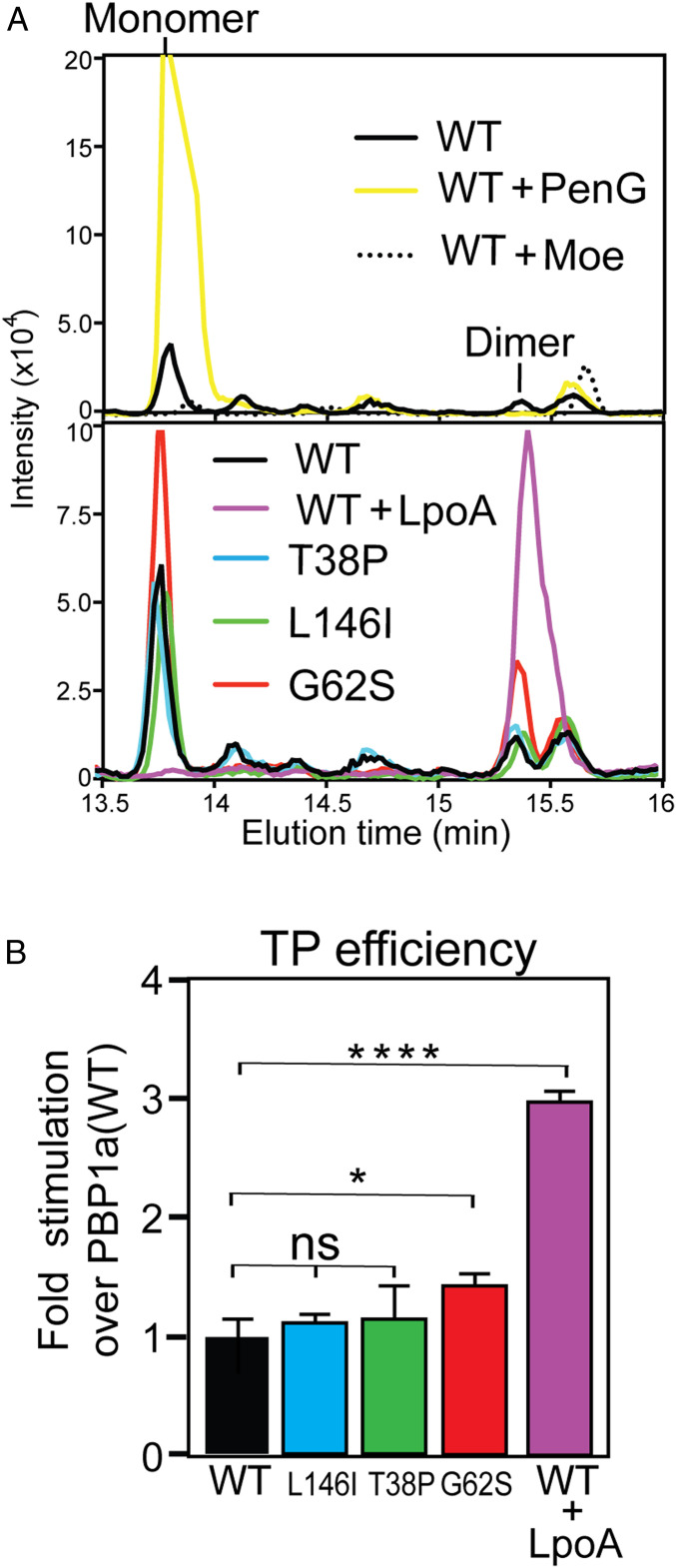
To test the TP activity of the PBP1a* variants, we used similar assays but with native E. coli lipid II substrate. In this case, the resulting PG products were digested with mutanolysin and subjected to liquid-chromatography–mass-spectrometry (LC-MS) to detect cross-linked muropeptide species. PBP1a(WT) generated LC-MS peaks corresponding to monomeric (uncross-linked) and dimeric (cross-linked) muropeptide products (Fig. 4A). The synthesis of both products was inhibited by the addition of the PGTase inhibitor moenomycin, as expected. Additionally, the production of dimeric products was blocked by penicillin G (penG) treatment. Notably, glycan polymerization was greatly stimulated by penG as evidenced by the large increase in the monomeric peak observed following drug treatment. This result is consistent with a previous study showing that in vitro PG polymerization by aPBPs is activated by β-lactams (31). Like PBP1a(WT), all of the PBP1a* variants produced both monomeric and dimeric products.
Fig. 4.
E. coli PBP1a* variants are not strongly activated for TPase function. (A) Shown are extracted ion LC-MS chromatograms of mutanolysin-treated products from PG synthesis reactions with E. coli lipid II (200 µM) and purified PBP1a(WT) or its PBP1a* derivatives (5 µM) incubated at 25 °C for 5 min prior to heat inactivation. The Upper panel compares representative traces from PBP1a(WT) without or with penG (50 µM) or moenomycin (50 µM) treatment as indicated. The Lower panel compares representative traces from reactions with PBP1a(WT) and its indicated PBP1a* derivatives as well as PBP1a(WT) with added LpoA (10 µM). All traces are averages of three to five independent reactions for each condition/protein variant. (B) The PG cross-linking efficiency in each of the reactions was calculated by dividing the amount of dimer formed by the total PG produced (monomer + dimer). Results are averages of three experiments. Error bars represent the SEM value. Unpaired Student’s t test was used to determine whether the responses of the cells to the treatment was significantly different (ns, not significant P > 0.05, *P ≤ 0.05, ****P ≤ 0.0001).
The cross-linking efficiency of each enzyme was assessed by determining the ratio of dimeric muropeptides produced relative to the total amount of PG synthesized. Of the PBP1a* variants, only PBP1a(G62S) showed a modest increase in TP activity relative to the wild-type enzyme (Fig. 4B). This increase was much lower than that observed when LpoA was added to PBP1a(WT) (Fig. 4B). Notably, only the PBP1a(G62S) variant produced more monomeric muropeptides than PBP1a(WT), as expected for an enzyme with activated PGTase activity. In order to efficiently detect PG cross-linking, much higher levels of the lipid II substrate (200 vs. 40 µM) were used in the reactions to measure TPase activity than in the PGTase assays above. Thus, mechanism of PGTase activation conferred by the amino acid changes in the PBP1a* variants may be at the level of substrate binding and therefore best observed at lower substrate concentrations. Further experiments will be required to test this possibility. Nevertheless, our overall biochemical analysis indicates that the primary effect of the amino acid changes in the characterized PBP1a* variants is PGTase activation with only minimal if any increase in TP activity.
The TP Activity of PBP1a(G62S) Is Essential for Its LpoA Bypass Function.
PBP1a has been found to interact with PBP2, the monofunctional bPBP with TP activity that is required for cell elongation (31). PBP2 has also been shown to be capable of cross-linking PBP1a products in vitro (31). We therefore wondered whether the high PGTase activity of the PBP1a* variants alone was sufficient for LpoA bypass activity with TP activity potentially being supplied by another TPase, like PBP2. To investigate this possibility, we tested the LpoA bypass activity of a PBP1a(G62S) variant with an additional substitution (S473A) that inactivates its TPase domain. Whereas PBP1a(G62S) supported the growth of MM20 [ΔlpoA Para::ponB] following PBP1b depletion, PBP1a(G62S, S473A) could not (SI Appendix, Fig. S1), indicating that TP activity remains required for this PBP1a* variant to function despite its highly elevated PGT activity. Thus, the activated PBP1a derivatives are likely to be directly incorporating the glycan strands they produce into the PG matrix, even though they may ultimately be aided by additional cross-linking enzymes like PBP2.
LpoA Is Required to Activate PG Polymerization by PBP1a In Vivo.
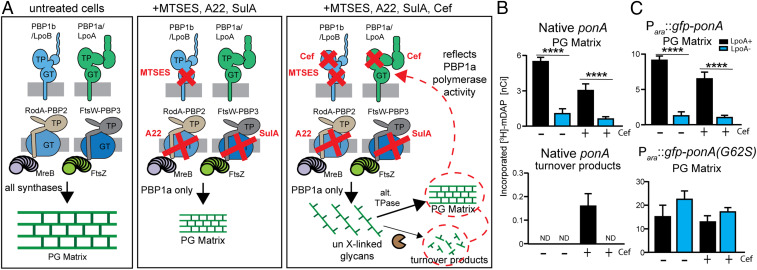
Based on the results from the biochemical analysis of the PBP1a* variants, we wondered whether LpoA might be critical for the activation of PG polymerization by PBP1a in vivo. To measure the PGTase activity of PBP1a in cells, we took advantage of a radiolabeling assay capable of monitoring PG polymerization by specific PG synthases in E. coli (25, 26). The assay is based on the observation that when a TPase is inactivated by a β-lactam, the uncross-linked PG polymers produced by its partner PGTase are rapidly degraded by lytic transglycosylases (26). Thus, measurement of the amount of PG turnover products produced following β-lactam treatment and radiolabeling with the PG-specific amino acid [3H]-mDAP provide an indirect readout of cellular PG polymerase activity. The assay can be tailored to monitor the activity of a particular PG synthase by combining the use of β-lactams that target specific TPases with genetic and chemical perturbations that inactivate all synthases, save for the one to be assayed (5, 25, 26). For our purposes, we expressed sulA to block PG synthesis by the SEDS-bPBP synthase FtsWI in the divisome and we treated cells with the compound A22 to block the activity of the corresponding RodA-PBP2 synthase of the elongation machinery (5, 25, 26). With both SEDS-bPBP synthases inactivated, only the aPBPs remain active, and their PG polymerase activity can be assessed following treatment with the aPBP-specific β-lactam cefsulodin (32). To further focus the assay on PBP1a, we employed a strain that produces PBP1b with an S247C substitution that allows its PGTase activity to be inhibited by treating cells with the cysteine-reactive compound 2-sulfonatoethyl methanethiosulfonate (MTSES) (5). Thus, in the presence of MTSES and cefsulodin, the radiolabeling assay can be used as a measure of the cellular PG polymerase activity of PBP1a (Fig. 5A).
Fig. 5.
PG synthesis by PBP1a requires LpoA in vivo. (A) Overview of the in vivo assay for PBP1a PG synthesis and PGTase activity. In untreated cells (Left), all four major PG synthases contribute to radiolabel incorporation into PG. In cells treated with MTSES, A22, and SulA (Center), PBP1a is the only active PG synthase such that radiolabel incorporation into the PG layer reflects the combination of its PGTase and TPase activities. Adding the β-lactam cefsulodin (Right) results in the inactivation of the TPase domains for both aPBPs. Only the PGTase activity of PBP1a remains active, leading to the production of uncross-linked strands, which largely appear to be cross-linked into PG by alternative TPase enzymes with a small amount of the glycan products being degraded by lytic transglycosylases to produce turnover products. Thus, measuring the level of PG synthesis in the presence of cefsulodin provides an indirect readout of the relative PGTase activity of PBP1a. (B) Cells of HC529(attHKHC859) [ΔlysA ΔampD ΔpbpC ΔmtgA ponB(S247C) (Ptac::sulA)] and its ΔlpoA derivative were treated with IPTG (1.5 mM) to induce SulA production followed by treatment with A22 (10 µg/mL), MTSES (1 mM), with or without additional treatment with cefsulodin (100 µg/mL), as indicated. After 5 min of treatment, cells were pulse-labeled with [3H]-mDAP for 10 min and label incorporation into the PG matrix and turnover products was assessed (Materials and Methods). Results are averages of three experiments. Error bars represent the SEM value. ND, not detectable. Note that the y axis scale for the turnover products is different from that for PG incorporation. Values for the detected turnover products are near background. (C) Cells of HC533(attHKHC859) [ΔlysA ΔampD ΔponA ΔpbpC ΔmtgA ponB(S247C) (Ptac::sulA)] and its ΔlpoA derivative with the indicated Para::gfp-ponA constructs were grown an analyzed as for B with the exception that arabinose was added to induce either GFP-PBP1a(WT) (Upper) or GFP-PBP1a(G62S) (Lower), as indicated. Unpaired Student’s t test was used to determine whether the responses of the cells to the treatment was significantly different (****P ≤ 0.0001).
In cells of the labeling strain HC529(attHKHC859) [ΔlysA ΔampD ΔpbpC ΔmtgA ponB(S247C) (Ptac::sulA)] that were inhibited for all PG synthases but PBP1a (+SulA, +A22, +MTSES, referred to as SAM-treated), the incorporation of radiolabel into PG was detected (Fig. 5B). Strikingly, however, when lpoA was deleted in the labeling strain, PG synthesis was largely inhibited in the SAM-treated cells (Fig. 5B). In the absence of cefsulodin, no PG turnover products were detected for either strain following SAM treatment, as expected (Fig. 5B). Unexpectedly, the addition of cefsulodin to the SAM-treatment regimen did not result in a strong reduction of new PG incorporation into the matrix in LpoA+ cells, nor did it result in a corresponding conversion of the signal into turnover products (Fig. 5B). Instead, only a minor reduction in PG synthesis was observed, with a small portion being converted to turnover products, which were only barely detectable over background. This result indicates that PG glycans made by PBP1a in the presence of cefsulodin can still be incorporated into the matrix to prevent their degradation. It is possible that this incorporation is mediated by residual PBP1a cross-linking activity that escapes inactivation by the drug. However, we think this is unlikely because the concentration of cefsulodin required to acylate the active sites of 50% of the cellular PBP1a molecules was previously measured to be 0.47 µg/mL (32) and our assays include a much higher dose of the β-lactam (100 µg/mL). We therefore think the glycans made by cefsulodin-targeted PBP1a are likely being incorporated into the matrix by other cross-linking enzymes, possibly PBP2 or one of the L,D-transpeptidases (LDTs) (9, 31). Although this alternative cross-linking activity prevents robust turnover of the PBP1a glycan products, the observed PG incorporation in the presence of cefsulodin nevertheless reflects the PGTase activity of PBP1a (Fig. 5A). This incorporation was again found to be LpoA-dependent (Fig. 5B), indicating that PG polymerization by PBP1a requires LpoA.
To further explore the LpoA-dependence of PBP1a in cells, we used a labeling strain HC533(attHKHC589) [ΔlysA ΔampD ΔponA ΔpbpC ΔmtgA ponB(S247C) (Ptac::sulA)] or its ΔlpoA derivative, both of which lack native PBP1a but produce either GFP-PBP1a(WT) or GFP-PBP1a(G62S) from an integrated, arabinose-inducible chromosomal construct. For cells producing GFP-PBP1a(WT), the results mirrored those from cells producing PBP1a from its native locus (Fig. 5C). The incorporation of nascent PG into the matrix was observed in SAM-treated cells with or without cefsulodin treatment when LpoA was functional but not when it was inactivated (Fig. 5C). In contrast, in SAM-treated cells producing GFP-PBP1a(G62S), nascent PG incorporation was observed in all samples whether or not LpoA was inactivated (Fig. 5C). Taken together, the results of the radiolabeling assays indicate that LpoA is required for PBP1a to synthesize glycan strands in cells and that the PBP1a* variants likely overcome this requirement by allowing the enzyme to spontaneously transition to the active, polymerization-competent state.
Discussion
The first set of activators required for PG synthesis by the aPBPs were the LpoA and LpoB proteins of E. coli (13, 14). Since then, orthologs have been demonstrated to serve a similar role in other gram-negative bacteria (15–17), and unrelated proteins with potentially analogous roles have been identified in gram-positive organisms (33, 34). Although the use of accessory proteins to promote the activity of the aPBPs appears to be widespread in bacteria, we are only just beginning to learn about the role these auxilliary factors play in the overall regulation of PG synthesis. What little we know is principally based on biochemical analysis of the effect of the Lpo proteins on aPBP activity in a purified system (13, 14, 18–20). While such studies have provided important clues toward understanding Lpo protein activity, they have not given us the full picture of the cellular function of these factors. Connecting the in vitro activity of the Lpo factors to their in vivo function has been especially difficult given that purified aPBPs are relatively active enzymes in vitro in the absence of activators, yet the same synthases appear to function poorly or not at all in cells when their cognate Lpo protein is inactivated. In this report, we used a combination of genetics, biochemistry, and physiological labeling to show that LpoA, the most widely conserved aPBP activator in gram-negative bacteria (14), has a much more profound role in controlling the enzymatic activity of its cognate PG synthase in cells than anticipated from prior biochemical analyses.
In a purified system, the major effect of LpoA on PBP1a is the stimulation of its TPase activity (14, 19). It also has a modest effect on the PGTase activity of the synthase, but this effect was previously found to be eliminated upon the inhibition of TPase activity by β-lactam treatment (19). It has therefore been generally accepted that the activation of TPase activity is the main function of LpoA (1). Our genetic results provided the first indication that LpoA may be doing more than affecting PG cross-linking by PBP1a in cells. The majority of the identified PBP1a* variants that bypass the LpoA requirement in both E. coli and P. aeruginosa enzymes had amino acid substitutions in the PGTase domain or the interdomain linker region of the synthase. When the E. coli derivatives were purified, they were found to have elevated PGTase and TPase activity, with PGTase activity being activated to a much greater degree. This result suggested that LpoA may be required to promote PG polymerization by PBP1a rather than just affecting its PG cross-linking activity. Indeed, when we monitored the PG polymerase activity of PBP1a in vivo using a radiolabeling assay, little to no activity was detected in cells lacking LpoA. In contrast, PG synthesis was detected for the PBP1a(G62S) variant in ΔlpoA cells. We therefore conclude that LpoA is required to promote PG polymerization by PBP1a in vivo. Importantly, biochemical results suggest that LpoA is probably also affecting the TPase activity of PBP1a in cells. However, because PG polymerization must precede cross-linking, it is therefore the more ideal target for the regulation of PG synthesis. Thus, we think that PGTase activation is likely to be the most critical cellular function of LpoA.
Notably, nascent PG incorporation into the matrix by PBP1a was detected in the radiolabeling assay even in the presence of cefsulodin, which inhibits the TP domain of this synthase (32). Thus, additional TP enzymes in the cell may partner with PBP1a to enhance the incorporation of its products into the mature wall. This potential support from alternative cross-linking enzymes is apparently not enough to fully substitute for the intrinsic cross-linking activity of PBP1a given that cefsulodin treatment is lethal and that the TPase inactivated variant PBP1a(G62S, S473A) was unable to bypass LpoA. Nevertheless, it will be interesting to determine the identity of the factor providing the additional cross-linking activity. The bPBP-type enzyme PBP2 is an excellent candidate based on the previous demonstration that is interacts with PBP1a and can cross-link its products in vitro (31). Further work will be required to test this possibility and also investigate the potential of other cross-linking enzymes like the LDTs to work with PBP1a to assemble the PG matrix.
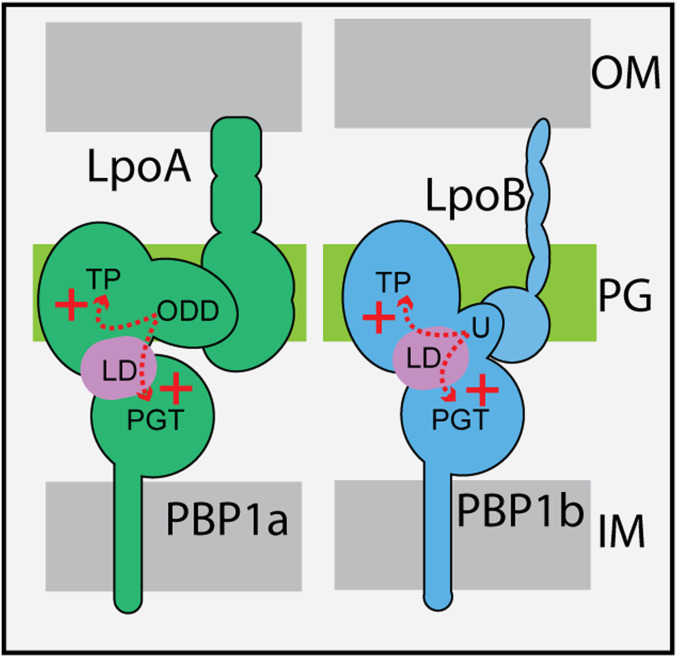
Both PBP1a and PBP1b possess accessory domains that have been found to be the binding sites for their cognate Lpo factor (14). In the case of PBP1b, the UB2H domain that binds LpoB precedes the PGT domain in the primary sequence but is positioned between the two catalytic domains in the folded structure (18, 35). Amino acid changes in the interdomain linker region of PBP1b located just behind the UB2H domain were previously found to stimulate the PGTase activity of PBP1b and allow it to function in cells lacking its activator (21). Binding of LpoB has also been found to cause structural changes in the UB2H domain that are likely to be propagated throughout PBP1b via the interdomain linker (36). It has therefore been proposed that LpoB activates the PGTase and TPase activities of PBP1b by inducing a conformational change in the interdomain region (21, 36). Accordingly, the amino acid changes in the LpoB bypass variants of PBP1b are thought to promote a similar change in the enzyme, allowing it to synthesize PG in cells in the absence of its activator (21). Our results with the PBP1a* variants suggest that, despite the dissimilarities between the enzymes and accessory proteins involved, the activation of PBP1a by LpoA may proceed via a mechanism similar to the activation of PBP1b by LpoB.
Instead of a UB2H domain, PBP1a has an accessory domain called the ODD (outer membrane PBP1a docking domain), which has an OB-fold (14, 37). In the primary sequence of the enzyme the ODD is found between the PGTase and TPase domains (14, 37). Rather than being topologically connected with the interdomain linker region like UB2H, the ODD is connected to the folded TPase domain (14, 37). Nevertheless, the OB-fold is spatially positioned between the PGTase and TPase domains and remains in relatively close proximity to the β-rich interdomain region like the UB2H domain of PBP1b (37). Two of the PBP1a* changes, G284D in P. aeruginosa and G62S in both the P. aeruginosa and E. coli enzymes, map to the interdomain region or its junction with the PGTase domain. With the exception of the W494G change in E. coli PBP1a, the other changes in the PBP1a* variants were found in the PGTase domain. These changes in the E. coli PBP1a* variants were found to significantly elevate the PGTase activity and modestly enhance TPase function, with the PBP1a(G62S) derivative being the most active on both counts. We therefore propose that PBP1a(G62S) and the other PBP1a* variants with changes in or near the PGTase domain are able to spontaneously adopt a conformation that mimics an activated state promoted by LpoA binding to the ODD. Moreover, the proximity of the ODD to the interdomain linker region of PBP1a and the location of the G62S substitution in this portion of the enzyme further suggests that, as proposed for PBP1b, the effects of LpoA binding to the ODD are likely to be transmitted to the PGTase domain through the interdomain linker. However, in this case the changes may either be transmitted directly or via changes in the conformation of the TPase domain that are then transmitted to the linker. The LpoA bypass activity of PBP1a(W494G) suggests the latter possibility, but unfortunately its in vitro activity could not be assessed biochemically due to technical difficulties with its purification. Regardless of whether the communication of the ODD domain with the linker domain is direct or indirect, our results support a model in which the mechanism of PBP1a activation by LpoA is similar to that of PBP1b activation by LpoB in that binding of the lipoprotein stimulates PGTase activity through changes in the interdomain linker (Fig. 6). Thus, this region of the aPBPs may generally be involved in regulatory mechanisms that modulate their enzymatic activity.
Fig. 6.
Model for aPBP activation by lipoprotein partners. Shown are schematics representing the structures of PBP1a and PBP1b in the cell envelope bound to their cognate lipoprotein partner. Genetic results suggest that both LpoA and LpoB activate their target synthase by directly or indirectly inducing changes in the interdomain linker region (lavender, linker domain, LD) which are then communicated to the enzymatic domains. Thus, even though the two aPBPs have different accessory domains and bind different partners, they may be activated by a similar mechanism.
In conclusion, our studies of PBP1a regulation have revealed that LpoA is not just involved in activating the TPase function of the enzyme as is commonly believed. It is also required to activate the upstream step of PG polymerization by the bifunctional synthase in cells. This finding indicates that the different types of outer membrane lipoprotein partners of aPBPs in gram-negative bacteria likely act similarly to promote the activity of their cognate synthases. Therefore, despite the unrelated activators and accessory domains involved, it may be possible to simultaneously target the activation of multiple different aPBPs in cells with a single small molecule as an alternative means of blocking PG synthesis for antibiotic development.
Materials and Methods
Media, Bacterial Strains, and Plasmids.
E. coli strains used were all derivatives of MG1655 and P. aeruginosa strains used were all derivatives of PAO1. Cultures were grown in lysogeny broth (LB), Terrific Broth (TB), M9 minimal medium containing 0.2% casamino acids and 0.2% sugar, or VBMM, as indicated. The following concentration of antibiotics were used: ampicillin (Amp), 50 µg/mL; chloramphenicol (Cam), 25 µg/mL; gentamicin (Gent), 15 µg/mL (E. coli); Gent, 30 µg/mL (P. aeruginosa); kanamycin (Kan), 25 µg/mL. The bacterial strains and plasmids used in this study are summarized in SI Appendix, Tables S1 and S2. Protocols for plasmid and strain construction can be found in SI Appendix. Regions of plasmids derived from PCR amplifications were verified by sequencing.
Selection for E. coli PBP1a* Variants.
A mutagenized ponA PCR product was generated using the ponA-containing plasmid pMM21 as a template and the primers pMM21-XbaI-PBP1a-F (5′-TCATCTAGACCGCGCGTTTG-3′) and PBP1a-HindIII-pMM21-R (5′-CGATAAGCTTTTGTCAGCAAACTG-3′). Two pools of mutagenized ponA PCR products were generated, one with the error-prone Pfu(D473G) polymerase and another with Taq polymerase. In each case, multiple PCR reactions were purified and combined to avoid jackpotting. The purified PCR libraries were digested with XbaI/HindIII and ligated into similarly digested pPR66 [cat lacIq Plac::empty] vector. Ligations were purified with the Qiagen PCR purification kit and electroporated into NEB 5-α electrocompetent E. coli cells (New England Biolabs). Libraries were plated on LB-Cam agar plates and grown overnight at 37 °C. The library was harvested by scraping colonies from the agar plates and suspending the cells in LB. Plasmid DNA was then purified from an aliquot of the resulting cell resuspensions using the Zymo Research Zyppy plasmid miniprep kit.
Each of the purified ponA plasmid libraries was transformed into electrocompetent MM20 [ΔlpoA Para::ponB] cells that were then plated on tryptone agar (1% tryptone, 0.5% NaCl) supplemented with 0.2% arabinose and Cam, and grown overnight at 37 °C. The resulting transformants were then scraped and resuspended as above to generate the final libraries used in the selections. Dilutions of the two libraries were plated on LB agar containing IPTG (30 or 250 µM) and no arabinose to select for plasmids encoding PBP1a* variants. Plasmids from surviving colonies were purified, confirmed to confer LpoA-bypass activity, and sequenced with Sanger sequencing.
Selection for P. aeruginosa PBP1a* Variants.
Overnight cultures of P. aeruginosa strain PA686 [ΔponB ΔlpoA] (17) were grown in LB without NaCl (LB0N) at 30 °C, before being pelleted, washed twice, and resuspended in an equal volume of VBMM minimal media. Cell suspensions were serially diluted in VBMM and then plated on VBMM agar plates at 30 °C for 2 d to select for spontaneous suppressors. Suppressors were purified under the same growth conditions. Overnight cultures of suppressors were grown in LB0N at 30 °C and genomic DNA was prepared using a Wizard Genomic DNA Purification Kit (Promega) and a Genomic DNA Clean & Concentrator-10 Kit (Zymo Research). The isolated genomic DNA was then prepared for sequencing using the NEBNext Ultra DNA Library Prep Kit for Illumina according to manufacturer’s instructions. The DNA concentration of the libraries was determined using the Qubit dsDNA HS Assay Kit and the size distribution of each library was determined using a High Sensitivity D1000 screen tape run on an Agilent 4200 TapeStation system. Sequencing was performed using a MiSeq Reagent Kit v3 and the Miseq System (Illumina). To confirm the LpoA bypass activity of the identified ponA alleles, they were amplified from genomic DNA of the suppressors and inserted into the pPSV38 vector for expression in strain PA686 [ΔponB ΔlpoA].
Protein Purification and In Vitro Enzyme Assays.
Detailed protocols for protein purification and enzyme assays are provided in SI Appendix. His6-SUMO-tagged derivatives of E. coli PBP1a or PBP1a* variants were expressed from pCB21 and its derivatives pMFS1, -4, -5, -6, and -19 in E. coli Rosetta(λDE3)/pLysSRARE, and His6-SUMO-LpoA(28-678) was expressed from plasmid pMM18 in Rosetta 2(DE3) cells. Protein expression and purification was carried out using a modified protocol based on that described previously (13). Lipid II substrates were purified as described previously (27), and the PGTase and TPase assays performed as described in the same study and related reports (27–30).
In Vivo PG Polymerase Assay.
The assay was performed essentially as described previously (5, 26). Cells of, MFS9(attHKHC859) [ΔlysA ΔampD ΔpbpC ΔmtgA ponB(S247C) (Ptac::sulA)], HC533(attHKHC859) [ΔlysA ΔampD ΔponA ΔpbpC ΔmtgA ponB(S247C) (Ptac::sulA)] or their ΔlpoA derivative were grown overnight in M9 medium supplemented with 0.4% (vol/vol) glycerol and 0.2% (wt/vol) casamino acids. The overnight cultures were diluted to OD600 = 0.04 in the same medium and were grown at 30 °C until their OD600 reached 0.25 to 0.3. Cell division was inhibited by inducing sulA expression with 1.5 mM IPTG and growth was continued for 30 min. In HC533(attHKHC859) strains harboring Para::gfp-ponA constructs, the overproduction of the PBP was also induced with the addition of 0.4% (vol/vol) arabinose during this time. The OD600 of each culture was then adjusted to 0.3, and cells were treated with A22 (10 µg/mL), MTSES (1 mM), and cefsulodin (100 µg/mL) as indicated. Growth was continued for 5 min before 1 μCi of [3H]-mDAP was added. Labeling proceeded for another 10 min. After labeling, cells were centrifuged in a tabletop centrifuge at maximum speed (21,000 × g) at 4 °C. The supernatant was removed, and the cell pellets were resuspended in 0.7 mL of cold (4 °C) HPLC grade water and placed on a heat block at 90 °C for 30 min to extract hydrophilic molecules from the cells (hot water extract). After the heat treatment, tubes were cooled on ice, and each solution was centrifuged at 200,000 × g for 20 min at 4 °C. The supernatant that contains the hydrophilic PG turnover intermediates was removed and lyophilized under vacuum overnight. PG turnover products were then separated and quantified as described in SI Appendix. The pellets were used to determine how much [3H]-mDAP was incorporated into the PG as described in the SI Appendix.
Supplementary Material
Acknowledgments
The authors thank all members of the T.G.B. and Rudner laboratories for advice and helpful discussions; and Suzanne Walker and members of her laboratory, especially Atsushi Taguchi, for their advice in setting up the PGTase and TPase assays. This work was supported by NIH Grants R01AI083365 (to T.G.B.) and F31AI122363 (to J.L.B.), and funds from the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2108894118/-/DCSupplemental.
Data Availability
All study data are included in the main text and SI Appendix.
References
- 1.Egan A. J. F., Errington J., Vollmer W., Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 18, 446–460 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Fisher J. F., Mobashery S., Constructing and deconstructing the bacterial cell wall. Protein Sci. 29, 629–646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P., The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Meeske A. J., et al., SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho H., et al., Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol. 1, 16172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohs P. D. A., et al., A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLoS Genet. 14, e1007726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taguchi A., et al., FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587–594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vigouroux A., et al., Class-A penicillin binding proteins do not contribute to cell shape but repair cell-wall defects. eLife 9, e51998 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morè N., et al., Peptidoglycan remodeling enables Escherichia coli to survive severe outer membrane assembly defect. MBio 10, e02729–e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straume D., et al., Class A PBPs have a distinct and unique role in the construction of the pneumococcal cell wall. Proc. Natl. Acad. Sci. U.S.A. 117, 6129–6138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yousif S. Y., Broome-Smith J. K., Spratt B. G., Lysis of Escherichia coli by beta-lactam antibiotics: Deletion analysis of the role of penicillin-binding proteins 1A and 1B. J. Gen. Microbiol. 131, 2839–2845 (1985). [DOI] [PubMed] [Google Scholar]
- 12.Kato J., Suzuki H., Hirota Y., Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol. Gen. Genet. 200, 272–277 (1985). [DOI] [PubMed] [Google Scholar]
- 13.Paradis-Bleau C., et al., Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143, 1110–1120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Typas A., et al., Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143, 1097–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dörr T., et al., A novel peptidoglycan binding protein crucial for PBP1A-mediated cell wall biogenesis in Vibrio cholerae. PLoS Genet. 10, e1004433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin J., Sun Y., Mao Y., Jin M., Gao H., PBP1a/LpoA but not PBP1b/LpoB are involved in regulation of the major β-lactamase gene blaA in Shewanella oneidensis. Antimicrob. Agents Chemother. 59, 3357–3364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greene N. G., Fumeaux C., Bernhardt T. G., Conserved mechanism of cell-wall synthase regulation revealed by the identification of a new PBP activator in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 115, 3150–3155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egan A. J. F., et al., Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc. Natl. Acad. Sci. U.S.A. 111, 8197–8202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupoli T. J., et al., Lipoprotein activators stimulate Escherichia coli penicillin-binding proteins by different mechanisms. J. Am. Chem. Soc. 136, 52–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caveney N. A., et al., Structure of the peptidoglycan synthase activator LpoP in Pseudomonas aeruginosa. Structure 28, 643–650.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markovski M., et al., Cofactor bypass variants reveal a conformational control mechanism governing cell wall polymerase activity. Proc. Natl. Acad. Sci. U.S.A. 113, 4788–4793 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sathiyamoorthy K., Vijayalakshmi J., Tirupati B., Fan L., Saper M. A., Structural analyses of the Haemophilus influenzae peptidoglycan synthase activator LpoA suggest multiple conformations in solution. J. Biol. Chem. 292, 17626–17642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vijayalakshmi J., Akerley B. J., Saper M. A., Structure of YraM, a protein essential for growth of Haemophilus influenzae. Proteins 73, 204–217 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Jean N. L., et al., Elongated structure of the outer-membrane activator of peptidoglycan synthesis LpoA: Implications for PBP1A stimulation. Structure 22, 1047–1054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uehara T., Park J. T., Growth of Escherichia coli: Significance of peptidoglycan degradation during elongation and septation. J. Bacteriol. 190, 3914–3922 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho H., Uehara T., Bernhardt T. G., Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiao Y., et al., Lipid II overproduction allows direct assay of transpeptidase inhibition by β-lactams. Nat. Chem. Biol. 13, 793–798 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsh M. A., et al., Identification of a functionally unique family of penicillin-binding proteins. J. Am. Chem. Soc. 139, 17727–17730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao Y., et al., Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J. Am. Chem. Soc. 136, 14678–14681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taguchi A., Kahne D., Walker S., Chemical tools to characterize peptidoglycan synthases. Curr. Opin. Chem. Biol. 53, 44–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banzhaf M., et al., Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol. Microbiol. 85, 179–194 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Curtis N. A., Orr D., Ross G. W., Boulton M. G., Affinities of penicillins and cephalosporins for the penicillin-binding proteins of Escherichia coli K-12 and their antibacterial activity. Antimicrob. Agents Chemother. 16, 533–539 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fenton A. K., et al., Phosphorylation-dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP. Proc. Natl. Acad. Sci. U.S.A. 115, 2812–2817 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenton A. K., El Mortaji L., Lau D. T. C., Rudner D. Z., Bernhardt T. G., CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat. Microbiol. 2, 16237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sung M.-T., et al., Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 8824–8829 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egan A. J. F., et al., Induced conformational changes activate the peptidoglycan synthase PBP1B. Mol. Microbiol. 110, 335–356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han S., et al., Distinctive attributes of β-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics. J. Am. Chem. Soc. 133, 20536–20545 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.