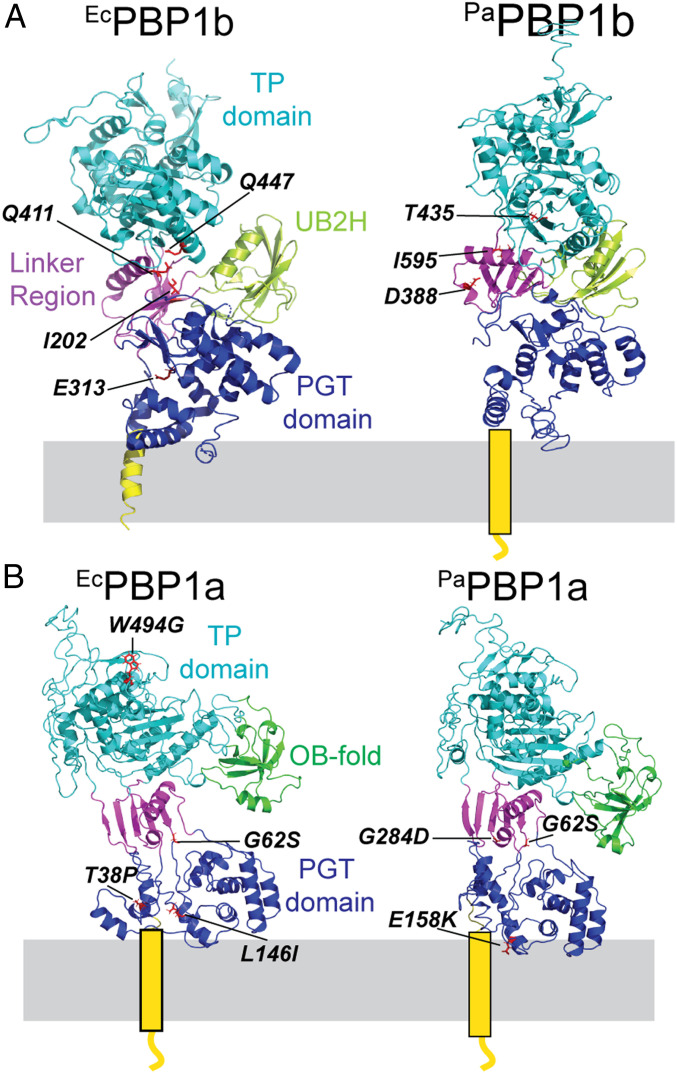
Fig. 1.
PBP structures and locations of activator bypass substitutions. Shown is the structure of E. coli PBP1b (PDB ID code 3VMA) (35) (A) and model structures of P. aeruginosa PBP1b (A), E. coli PBP1a (B), and P. aeruginosa PBP1a (B) made using i-Tasser. The PBP1a structures were based on the structure from Acinetobacter baumannii (PDB ID code 3UDF) (37). The different domains of the PBPs are color-coded. The locations of the previously identified LpoB/LpoP bypass mutations in PBP1b (17) (A) are indicated on the structures as are the LpoA bypass substitutions in PBP1a identified in this report (B).