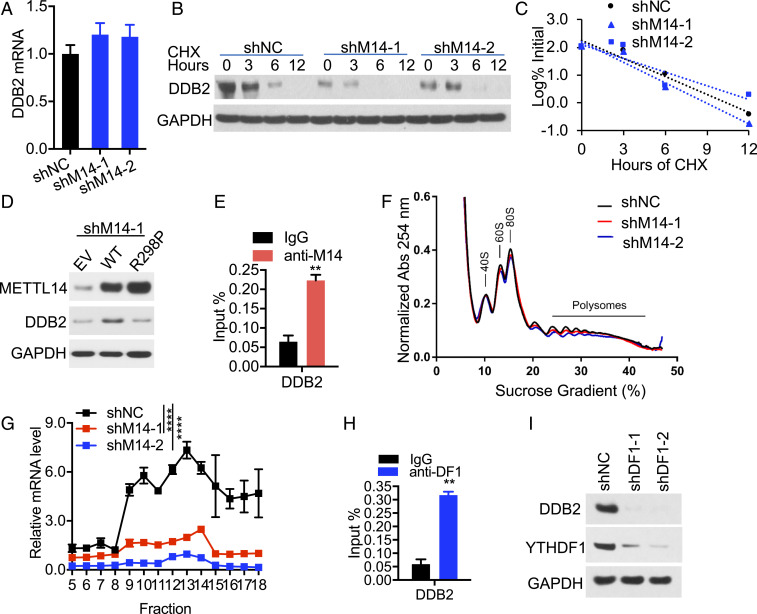
Fig. 5.
METTL14 regulates DDB2 translation through YTHDF1. (A) qPCR analysis of DDB2 mRNA levels in GAPDH in shNC or shM14-1/2 HaCaT cells (mean ± SD, n = 6). (B) Immunoblot analysis of DDB2 and GAPDH in shNC or shM14-1/2 HaCaT cells treated with or without cycloheximide (CHX, 100 μg/mL) over a time course. (C) Quantification of B. (D) Immunoblot analysis of METTL14, DDB2, and GAPDH in HaCaT cells transfected with empty vector (EV), WT, or R298P mutant METTL14. (E) CLIP-qPCR analysis for binding of METTL14 to the DDB2 transcript (mean ± SD, n = 3). (F and G) Polysome profiling assays in fractionations of HaCaT cell lysates (F) and qPCR analysis of DDB2 mRNA levels in different fractions of ribosomes (G, mean ± SD, n = 3). (H) CLIP-qPCR analysis for binding of YTHDF1 to the DDB2 transcript (mean ± SD, n = 3). (I) Immunoblot analysis of DDB2 and YTHDF1 in shNC or shDF1-1/2 HaCaT cells. **P < 0.01; ****P < 0.0001; Student’s t test, significant differences from IgG (E and H) or shNC groups (G).