Fig. 5.
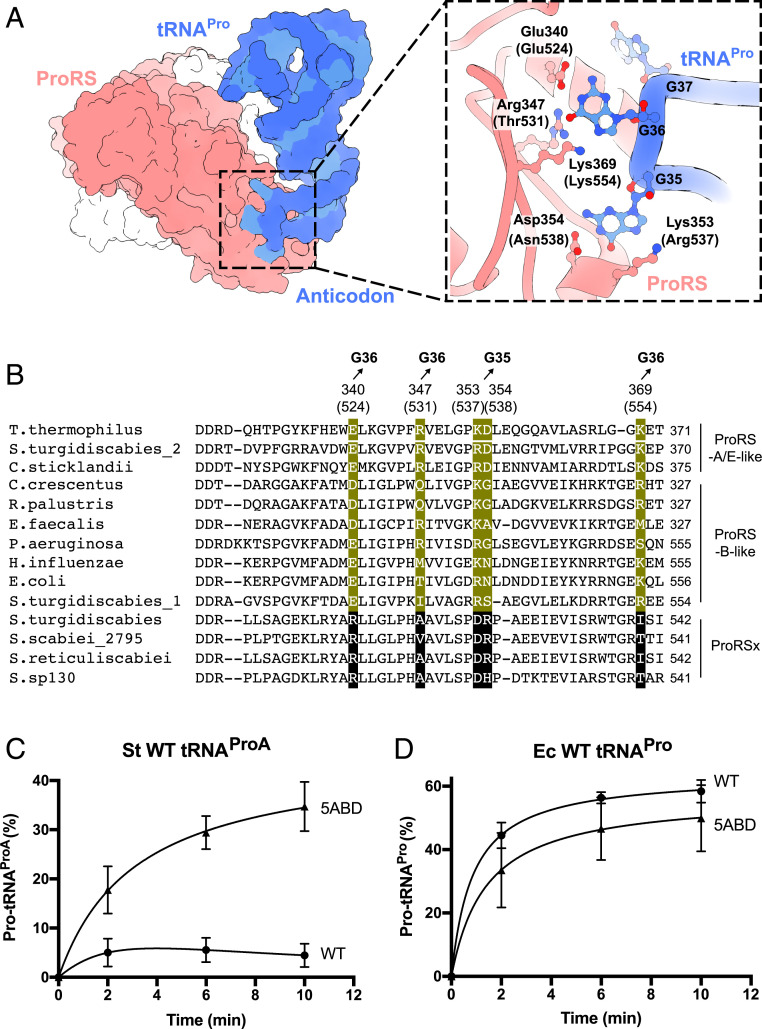
E. coli ProRS can be converted into a ProRSx-like enzyme by “transplanting” anticodon-binding residues from S. turgidiscabies ProRSx. (A) The crystal structure of T. thermophilus ProRS in complex with tRNAPro shows how ProRS recognizes the tRNAPro anticodon. A close-up view illustrates that the anticodon is recognized by five ProRS residues that bind the tRNAPro bases G36 and G35. Residue numbers in parentheses correspond to E. coli ProRS. (B) Multiple sequence alignment-comparing, anticodon-binding residues in canonical ProRS variants (B-type: bacterial-type and A/E-type: archaeal/eukaryotic-type) and the ProRSx variant. Residues that interact with the anticodon bases G35 and G36 of tRNAPro are highlighted in green. The corresponding residues in ProRSx are highlighted in black. Aminoacylation assays of S. turgidiscabies tRNAProA (C) and E. coli tRNAPro (D) with proline by either the WT E. coli ProRS (ProRS-Ec-WT) or its 5ABD variant (ProRS-Ec-5ABD). The results represent the average of three independent trials with the SD indicated.