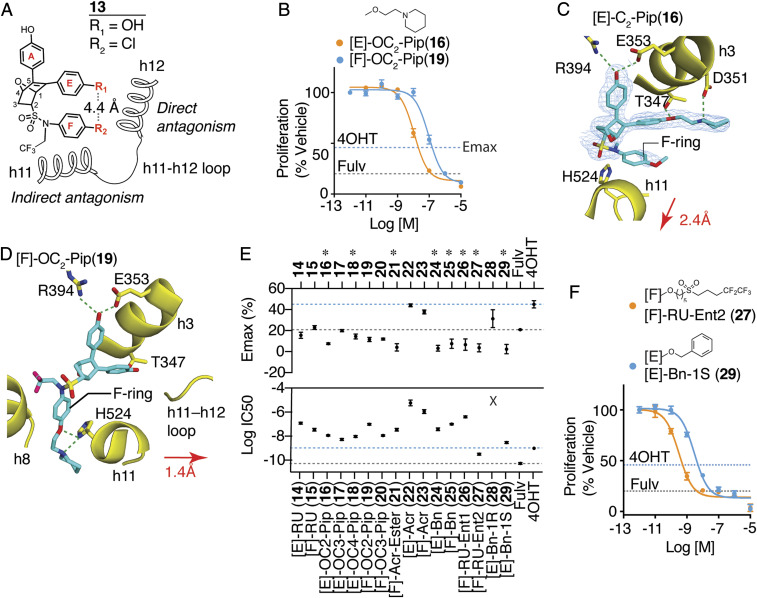
Fig. 1.
Dual-mechanism ER inhibitors fully suppress breast cancer cell proliferation. (A) Chemical structure of the OBHS-N scaffold and the orientation of substituents R1 and R2, with respect to h11 and h12 in the ER LBD (when R1 has a substituent, R2 is −OCH3 group; when R2 has a substituent, R1 is −OH for compounds 14–29; SI Appendix, Fig. S2 A–H). (B) Proliferation of MCF-7 cells treated for 5 d with 4OHT, fulvestrant (Fulv), or the indicated compounds. Datapoints are mean ± SEM, N = 6. M, Molarity. The horizontal lines indicate the Emax for 4OHT and fulvestrant (Fulv). (C) The structure of [E]-OC2-Pip (16)-bound ER LBD showed that the E-ring substituted piperidine H-bonding to Asp351 in helix 3 (h3), while the F-ring shifts helix 11 (h11) by 2.4 Å compared to an agonist bound structure. 2Fo-Fc electron density map contoured to 0.9 σ within 2 Å of the ligand. (D) The structure of [F]-OC2-Pip (19)–bound ER LBD shows that its [F]-OC2-Pip side chain exits the ligand-binding pocket between h8 and h11, H-bonds to His524, and shifts h11 toward h12. (E) Summary of dose–response curves for compound inhibition of proliferation of MCF-7 cells, shown in SI Appendix, Fig. S2 A–H. Datapoints are mean ± SEM, n = 6. * indicates pAdj < 0.05 (one-way ANOVA) for compounds with Emax > fulvestrant. (F) Selected dose curves from E. The lines indicate the Emax for 4OHT and Fulv.