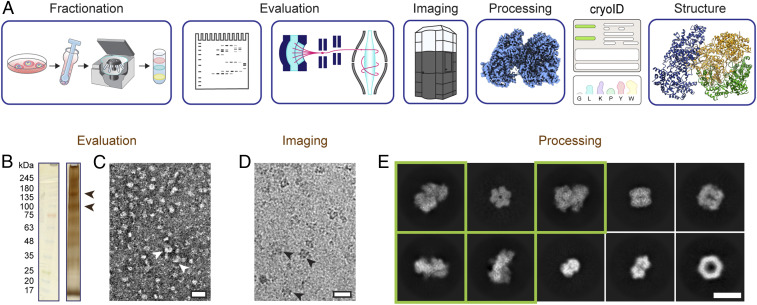
Fig. 1.
Endogenous structural proteomics workflow using cryoID. (A) Depiction of the workflow. P. falciparum parasites and parasitophorous vacuoles were saponin-released from P. falciparum–infected red blood cells, and the resulting parasite and vacuole pellets were lysed and fractionated using a sucrose gradient. The fractions were evaluated by SDS-PAGE, mass spectrometry, and negative stain EM. Cryo-EM imaging and analysis of the selected fraction yielded a 3.7-Å resolution cryo-EM density map. The proteins in the cryo-EM map were identified using cryoID and then modeled de novo, yielding the final atomic resolution structure of the RhopH complex. (B) Silver-stained SDS-PAGE of the selected fraction of the lysate. Arrowheads denote bands consistent with the molecular weights of the components of the RhopH complex. (C and D) Representative negative stain (C) and cryo-EM (D) micrographs of the selected fraction. (Scale bars, 20 nm.) Arrowheads denote particles consistent with 2D class averages of the RhopH complex. (E) Representative 2D class averages corresponding to multiple protein complexes present in the single dataset of cryo-EM micrographs. Class averages that ultimately gave rise to the structure of the RhopH complex presented here are boxed in green. (Scale bar, 10 nm.)