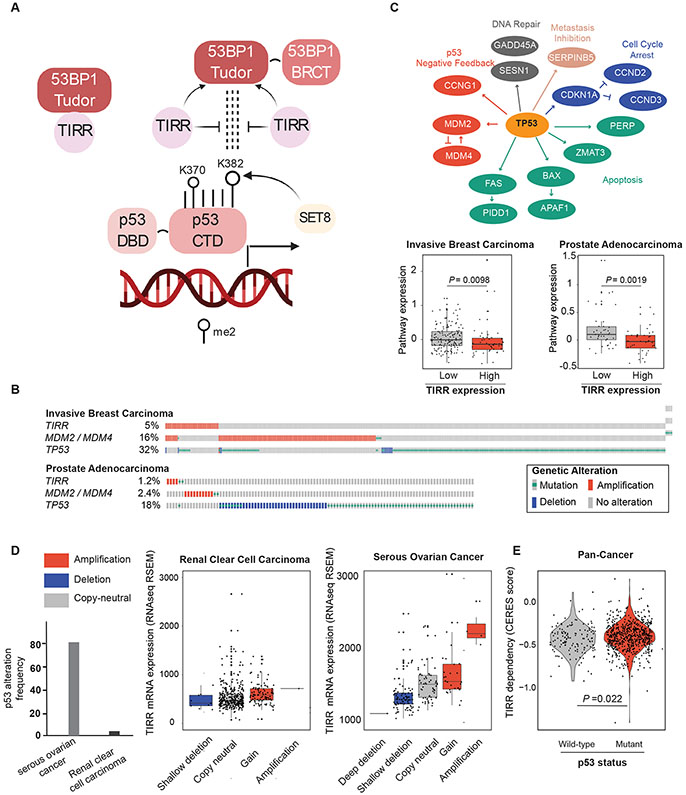
Figure 5: TIRR negatively regulates p53 signaling in cancer.
A) Model depicting TIRR as an upstream regulator of the 53BP1-p53 axis involved in p53 transactivation. The Tudor domain of 53BP1 interacts and with p53K382me2, a PTM of p53 that is associated with higher transactivation of p53. TIRR competes with dimethylated p53 for the Tudor domain of 53BP1.
B) OncoPrint showing the frequency at which TIRR, MDM2 / MDM4 and TP53 are altered in invasive breast carcinoma and prostate adenocarcinoma cohorts from TCGA (Cancer Genome Atlas, 2012, Hoadley et al., 2018). For visualization purposes, only samples with genetic alterations are shown in the plot. Putative driver mutations were chosen based on OncoKB driver annotations and copy number alterations were selected on GISTIC calls. OncoPrints were generated in the cBioPortal for Cancer Genomics [cbioportal.org].
C) Genes belonging to the p53 pathway for which expression data were used to calculate pathway expression estimates. Pathway expression in invasive breast carcinoma (top) and prostate adenocarcinoma (bottom) samples with and without TIRR amplification. Pathway expression scores were calculated as the average mRNA expression of p53 pathway members. RNAseq data for each p53 pathway gene were obtained from TCGA and were evaluated as z-scores relative to diploid samples. P-values, Wilcoxon test.
D) The level at which loss of TIRR is tolerated in human cancers is dependent on p53 status. Cancers with high frequencies of p53 alterations (serous ovarian cancer, right) can tolerate changes in TIRR expression through genomic loss, as a compensating mechanism for p53 activity. On the other hand, cancers with low frequency of p53 alterations (renal cancer) do not tolerate changes in TIRR expression that cause changes in p53 activity. Copy number events have been colored to indicate losses (blue) or gains (red) of genomic material. Gene expression (z-scores from RNAseq V2 RSEM) and copy number data were downloaded from TCGA (Cancer Genome Atlas, 2012).
E) Comparison of TIRR dependency scores (CERES) between 136 p53 WT and 633 p53 mutant cancer cell lines. p53 WT cell lines have a significantly higher dependency on TIRR. Dependency scores were calculated using DepMap’s Achilles Avana 20Q2 public release. [https://portals.broadinstitute.org/ccle]