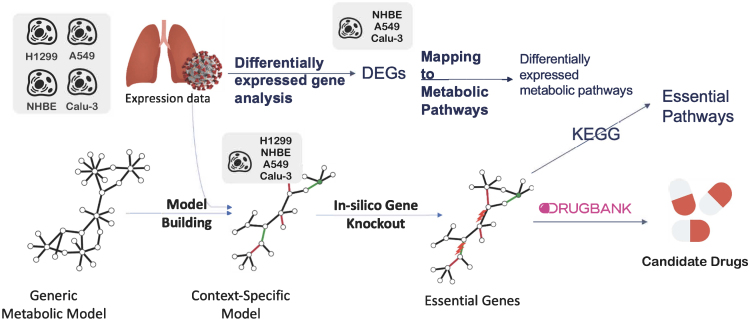
Figure 1.
Overview of the pipeline of essential gene prediction for SARS-CoV-2-infected lung cells
The viral biomass function (VBOF) was added to the generic metabolic models (Recon2.04 and Recon3D_01) (related to STAR Methods A.2.1) to build the infected generic models. Consistent versions of both the control and the infected generic models were obtained. Mock and infected lung expression data were used to build the context-specific models using rFASTCORMICS and the consistent control models as an input for the mock-specific models and the consistent infected models for infected-specific models, respectively. The objective functions were adjusted (related to STAR Methods A.2.2). Then, essential genes and gene pairs were identified by in silico gene knockout, before mapping to DrugBank V5 for drug repositioning to drugs and drug combinations.