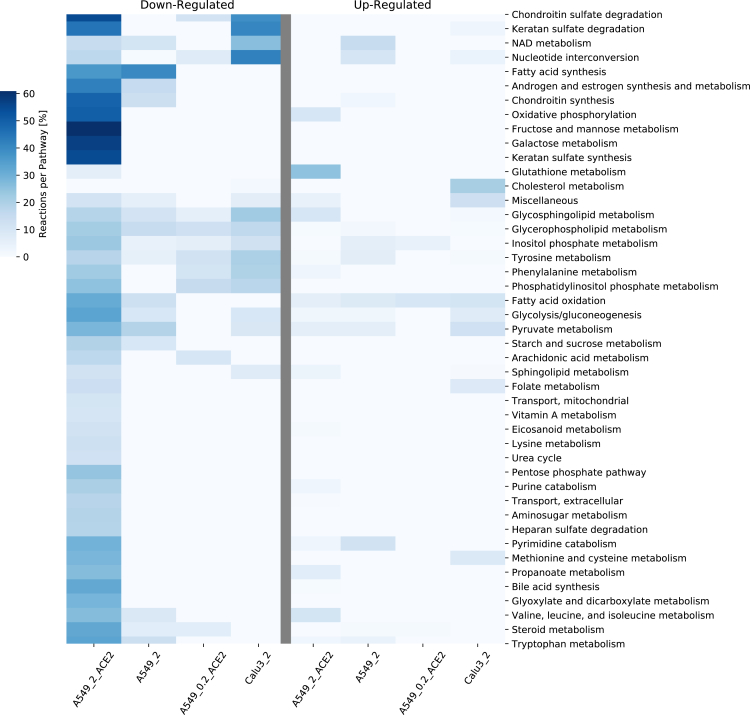
Figure 2.
Reactions per pathway Heatmap for pathway analysis of differentially expressed genes in the severity study
Differentially expressed genes (DEGs) were computed with DESeq2 (absolute log2 fold change >1, adjusted p-value < 0.05). The Down- and up-regulated were mapped to the pathways (subSystems) of Recon3D_01. The number of up and down-regulated reactions was then summed up to identify the top altered pathways in the infected lung cell lines in the severity study (related to STAR Methods A.1.2). The color code “Reactions per Pathway [%]” represents the number of enriched metabolic reactions in a pathway divided by the overall number of reactions in this pathway. The transfection of ACE2 at an MOI of two in the A549 cell lines caused the downregulation of many pathways, which was not seen when the MOI was decreased by a factor of ten.