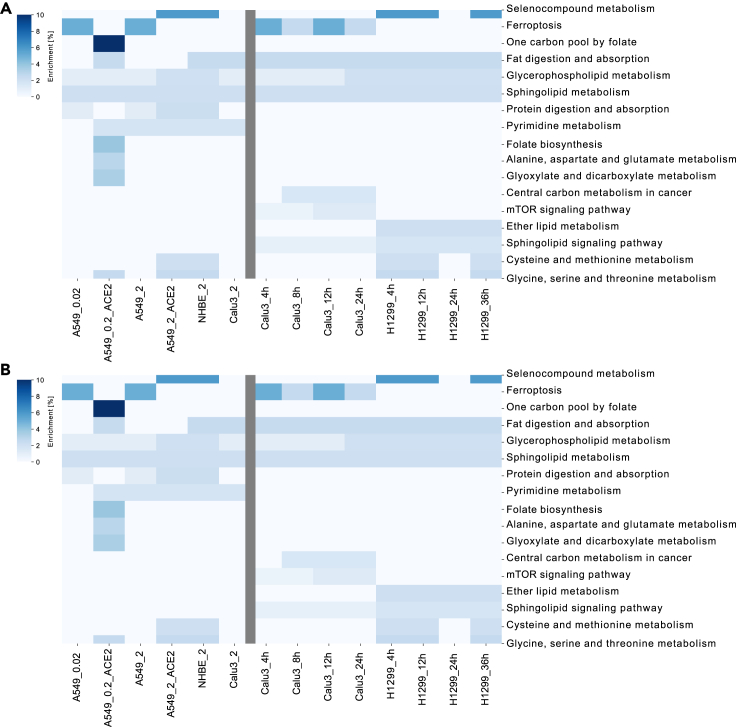
Figure 4.
Pathway analysis of the essential genes in the two lung studies
Identification of the pathways harboring the identified essential genes using Enrichr and Recon3D_01 subSystem, and hence the most critical pathways for the viral survival and proliferation across cell type, severity level, and time after infection. On the x axis from the left to the right, are the conditions of both the severity study and the time-series lung studies, respectively (related to STAR Methods B). Conditions in the severity study were named as cell line + ACE2 vector (if exists) + MOI. Conditions in the second lung cell line were named as cell line + time point.
(A) Enrichr enrichment: The color code in (A) "Enrichment [%]" represents the number of enriched genes in this pathway divided by the overall number of genes in this pathway.
(B) Metabolic pathway analysis: The color code in (B) "Reactions per Pathway [%]" represents the number of enriched metabolic reactions in a pathway divided by the overall number of reactions in this pathway.