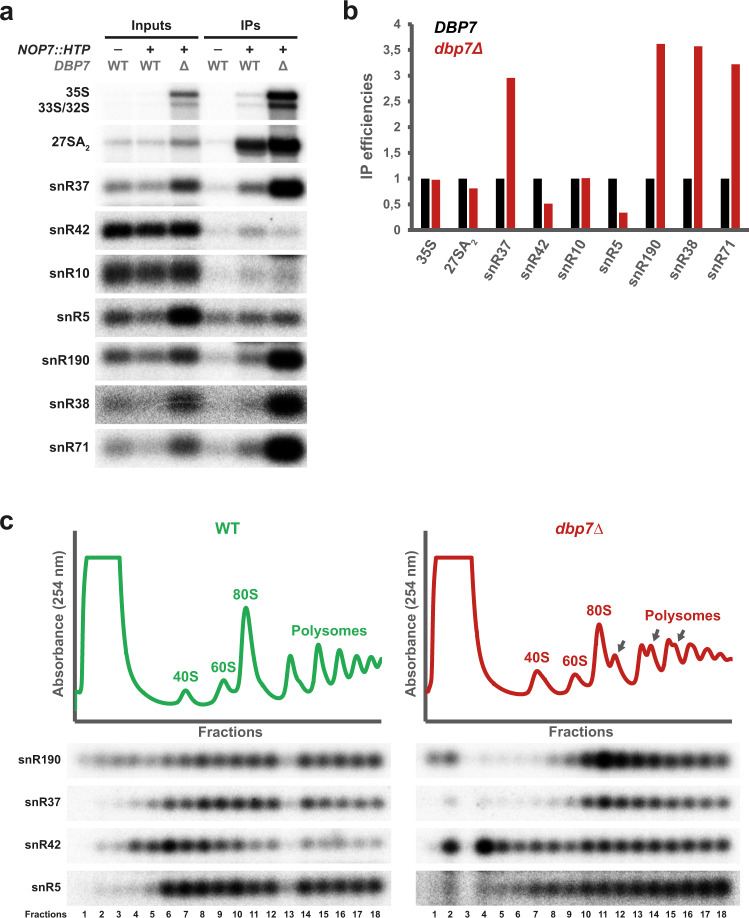
Fig. 5. snR190 is retained into preribosomal particles in the absence of Dbp7.
a Immunoprecipitation of Nop7-HTP-containing preribosomal particles in the presence or absence of Dbp7. Particles were immunoprecipitated from total cell extracts prepared from wild-type or dbp7Δ strains expressing an HTP-tagged version of Nop7. As a background control, the wild-type strain expressing untagged Nop7 was similarly processed. Some rRNA precursors and snoRNAs (indicated on the left) present in the total extracts (Inputs) or in the immuno-precipitates (IPs) were analyzed by northern blotting using selected radiolabeled probes (Supplementary Table 4). b Quantification of the northern blot signals obtained for the hybridizations shown in a using PhosphorImager data and the MultiGauge software. The histogram represents the co-immunoprecipitation efficiencies (IPs over Inputs) of the indicated RNA species with Nop7-HTP in the presence (DBP7, black) or absence (dbp7Δ, red) of Dbp7. c Sedimentation profile of snR190, snR37, snR42, and snR5 snoRNAs in the presence or absence of Dbp7. Total cellular extracts prepared from the wild-type (WT, green) or dbp7Δ (dbp7Δ, red) strains were centrifuged through 10–50% sucrose gradients. A254 was measured during gradient fractionation and the profiles are represented. RNAs extracted from the first 18 fractions were analyzed by northern blotting to detect the indicated snoRNAs. Note: the RNA samples corresponding to each gradient were processed separately (electrophoresis, transfer, hybridization, exposure, data acquisition). Therefore, the exposures presented for each snoRNA in the two strains have been chosen arbitrarily. Please refer to the intraseries quantifications (preribosome-bound versus free ratios) presented in Supplementary Fig. 10 for the interpretation of these data. This experiment has been performed once with the dbp7Δ strain and once with the strain expressing the Dbp7K197A catalytic mutant (Fig. 6c) with similar results for the snoRNAs tested.