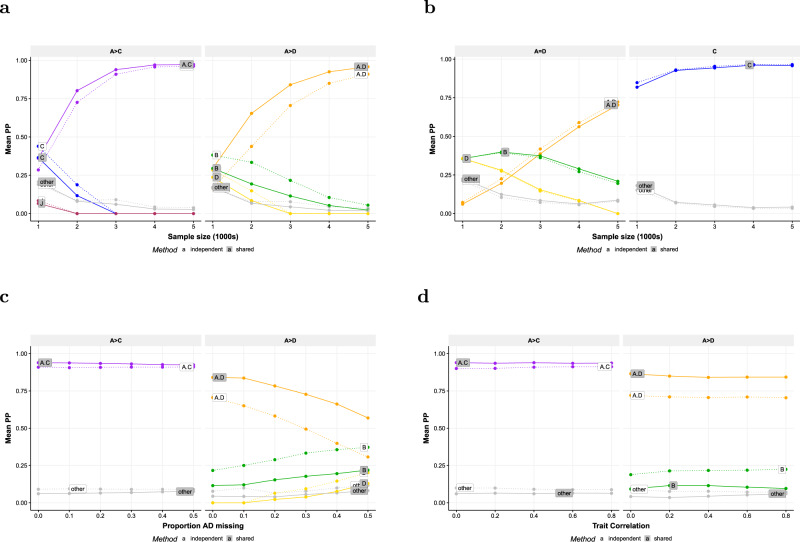
Fig. 2. Comparison of fine-mapping from flashfm and single-trait analyses.
When traits share a causal variant, flashfm has higher accuracy than single-trait finemapping, regardless of amount of missing data and trait correlation; both methods have similar accuracy when there are no shared causal variants. Causal variants were simulated for two traits with models defined by SNP groups from the IL2RA region. We vary sample size when the traits share a causal variant (a) and do not share any causal variants (b). At fixed sample size N = 3000, we vary the proportion of missing data for one trait (c) and vary the trait correlation (d). In a, c and d Trait 1 has causal variants A+C, while trait 2 has A+D causal variants, both A causal variants with the same effect size: βA = log(1.4) and βD = βC = log(1.25). In a and b there are no missing data and the sample size varies from 1000 to 5000. In c the sample size is fixed at 3000 and the proportion of missing data for trait A+D varies from 0 to 0.5. In d the sample size is fixed as 3000 and the correlation between traits varies. In b Trait 1 has causal variants A+D with βA = log(1.25) and βD = (1.25), while trait 2 has a single causal variant C with βC = log(1.25). Results are based on 300 replications. Source data are provided in Supplementary Data 1, Supplementary Data 1.2, 1.3, 1.6, 1.7.