Bonora et al.1 reported three families with subjects affected by a mitochondrial neurogastrointestinal encephalopathy (MNGIE) phenotype due to recessive mutations of LIG3, encoding ligase III, an enzyme present in both nucleus and mitochondria. A common molecular feature was reduced amount of mitochondrial DNA (i.e. mtDNA depletion). Affected subjects showed a quite variable severity, highlighted by the different age of symptom onset ranging from first months of age, until childhood/youth.
We describe here two baby siblings (Patients S1 and S2) harbouring biallelic variants in LIG3, with neonatal onset of a rapidly fatal myopathy, thus expanding the spectrum of this new genetic disease. One of the variants in these new cases selectively affects the mitochondrial isoform of ligase III, preserving the nuclear isoform, indicating that failure to maintain physiological mtDNA plays a key role in this disease.
The two babies belonged to a non-consanguineous Italian family (Fig. 1A). Patient S1 presented at birth with severe hypotonia, cardiorespiratory distress (Apgar scores: 1-3), and was immediately intubated. She had very high levels of lactate in plasma (6.1 mM, normal values 0.7–2.1), CSF (7.3 mM, normal value <2.1) and urine (2400 mMol/mol creatine, normal value <35). The EMG revealed myopathic features, while the EEG showed disorganized electric activity, possibly secondary to hypoxia. Despite admission to the NICU, she died after 17 days. An autoptic muscle biopsy showed numerous lipid vacuoles, a prevalence of hypotrophic fibres, and severe deficiency in cytochrome c oxidase staining, suggesting a mitochondrial myopathy. Biochemical activity of the oxidative phosphorylation (OXPHOS) complexes (C) in muscle show defects of C-I, III, IV and V (<5%, 9%, <5% and 32% of controls, respectively). In fibroblasts, only the activity of C-V was reduced (45%). The third child of the family (Patient S2), born at 36 weeks of gestation, presented at birth with a serious condition, leading to a fatal outcome within hours. Biochemical study on amniocytes showed C-IV decreased activity (56% of the control mean). No additional material was available from this baby.
Figure 1.
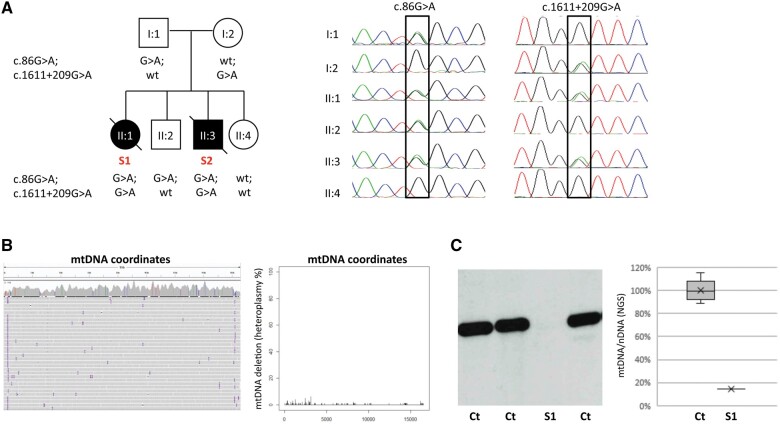
Genetic analysis of LIG3 and mitochondrial DNA. (A) Family pedigree of subjects 1 and 2 (Patients S1 and S2), with the sequencing chromatograms depicting the genomic regions corresponding to the exon 1 c.86G>A and the intronic c.1611+209G>A variants (NM_013975.4), in different family members. The two LIG3 variants segregated in the family. (B) Evaluations of mtDNA by next generation sequencing (NGS) in Patient S1 muscle DNA, as previously described by Legati et al.10: IGV snapshot (left) and NGS reads quantification (right) did not show any evidence of multiple mtDNA deletions. (C) Southern blot analysis (left) and a graph reporting the ratio between reads aligned to mtDNA and nuclear DNA (right), in muscle from Patient S1 and controls (Ct). A marked reduction in mtDNA amount in Patient S1 compared to control mean was evident.
Analysis of DNA extracted from Patient S1 muscle showed no multiple mtDNA deletions but marked mtDNA depletion (Fig. 1B and C). Hence, a diagnosis of mtDNA depletion syndrome (MDS) was made. MDS is an early-onset autosomal recessive disorder. To date, MDS pathogenic variants were found in diverse nuclear genes, including genes encoding enzymes of the mtDNA replication machinery, as well as genes encoding proteins controlling balanced mitochondrial nucleotide pool.2
Whole-exome sequencing in Patient S1 revealed the presence of a heterozygous nonsense variant in LIG3 (NM_013975.4:c.[86G>A], p.[W29*]), as the best candidate according to the patient’s phenotype. The variant was confirmed in Patient S2 DNA extracted from amniocytes, and also in the father and in one unaffected sibling, thus excluding a de novo dominant mutation (Fig. 1A). By means of a multi-omics approach performed on Patient S1 fibroblasts, we recently identified the second LIG3 variant in Patient S1 (Individual OM91786 in Kopajtich et al.3), compatible with a recessive trait; RNA sequencing showed aberrant splicing of exon 9 and a further whole genome sequencing identified one extremely rare heterozygous deep intronic variant (c.[1611+209G>A]). LIG3 transcript expression was reduced in RNA sequencing data5 and by quantitative PCR; accordingly, proteomics studies showed a residual amount of 56% for the LIG3 protein.3 In Patient S1 fibroblasts treated with ethidium bromide, mutant cells showed impaired recovery of induced mtDNA depletion,3 as also reported by Bonora et al.1 for other LIG3-mutant fibroblasts. Finally, genetic screening in the family confirmed segregation, with Patient S2 carrying both variants, and the intronic variant present in the mother and absent in the two healthy siblings (Fig. 1A).
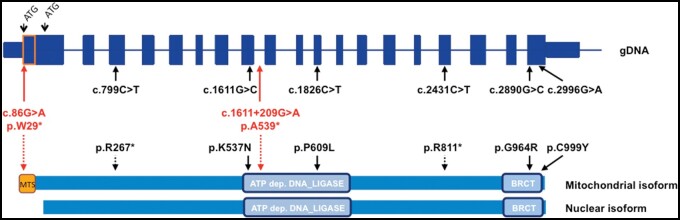
The human LIG3 gene contains two putative starting codons; the upstream ATG is the translation initiation site for the mitochondrial isoform (Fig. 2). The DNA sequence between the two ATGs encodes an amphipathic helix, which resembles already known mitochondrial targeting signal (MTS) peptides.5 Notably, the variant p.W29* identified in our family is located in the MTS; hence, the creation and translation of a transcript may still start from the second ATG, thus allowing the synthesis of the nuclear isoform, despite the presence of an early stop codon. Accordingly, proteomics data showed that half of the LIG3 protein is left, likely reflecting the nuclear isoform translated from the allele with the nonsense variant. In contrast, the second allele with the splice defect impairs both isoforms.
Figure 2.
Schematic picture of LIG3 and reported pathogenic variants. Schematic structure of the LIG3 gene and of the corresponding protein with both mitochondrial and nuclear isoforms, starting from two alternative ATGs. Variants identified in our family are in red, variants described by Bonora et al.1 are in black. Arrows indicate the positions in genomic DNA (gDNA) or protein sequence. Light blue boxes are two domains of ligase III where missense variants are located: the ATP dependent DNA ligase and BRCT (BRCA1 C-terminus) domains. Dotted lines indicate nonsense variants. MTS = mitochondrial targeting sequence.
Clinical and molecular features, in particular mtDNA depletion, of all the patients with LIG3 pathogenic variants fit well with the proposed role of ligase III. Indeed, despite DNA ligases playing a central role in nuclear DNA replication and repair, many studies have highlighted the involvement and requirement of ligase III in the maintenance of mtDNA integrity. MDS phenotypes are heterogeneous and usually classified as myopathic, encephalomyopathic, hepatocerebral or neurogastrointestinal.2,6 The latter is the only one with a later onset, in adolescence or early adulthood. Affected subjects from two families described by Bonora et al.1 presented the first symptoms as teenagers. Patients from the third family had an infantile onset (both at 2 months of age), with a fatal outcome at 2 years for one while the other was still alive at 3 years of age. All patients were characterized by chronic intestinal pseudo-obstruction and neurological involvement, with leukoencepalopathy, epilepsy and stroke-like episodes.
Our two siblings had a more severe form of myopathic MDS, leading to neonatal death. In contrast to the other patients, in our cases the EEG alterations were probably secondary to respiratory distress due to muscle weakness and hypotonia, as they had never been able to breathe autonomously. Moreover, no signs of gastrointestinal dysmotility were reported.
Considering all the reported LIG3 mutant patients (Fig. 2), it is difficult to identify easy genotype/phenotype correlations. Our patients showed the most severe phenotype, despite the presence of a mutation affecting only the mitochondrial isoform. This finding indicates that the specific presentation in our cases was mainly due to impairment of the mitochondrial isoform of ligase III rather than to defects in the nuclear or both isoforms.
The less severe subjects carried variants affecting both mitochondrial and nuclear LIG3 isoforms: compound heterozygosity for a nonsense (or predicted nonsense) variant and a missense change in the carboxyl-terminal domain was present in the subjects with milder phenotypes (Families 1 and 2 in Bonora et al.1); the presence of an allele in which the DNA ligase domain is preserved could be a possible explanation for the milder phenotype. In contrast, Family 3 harboured a missense variant in the DNA ligase domain, together with a premature stop codon variant. Immunoblot analysis of three patients’ cells (from the three families) showed a marked and similar decrease in LIG3 protein levels, but the quality/specificity of the used antibody were questionable, and the minimum detection limit was not assessed. A residual amount of functional protein (below the detection threshold) could explain the diverse clinical presentation and severity of the LIG3-related diseases.
In mitochondria, ligase III plays a number of different functions, including the crucial joining of the 5′ and 3′ termini of the nascent strands as a conclusive event of mtDNA replication and base-excision repair (BER). Germline LIG3 deletion is early embryonic lethal in mice.7 Conversely, a 90% knockdown of LIG3 is sufficient to maintain the normal copy number of mtDNA in HeLa cells, although these cells are unable to respond to exposure to DNA-damaging agents. It is thus possible that the normal levels of LIG3 in cells are apparently excessive to efficiently cope with various mtDNA injuries, such as oxidative damages.8 However, in the case of reduced amount of LIG3, mtDNA damages that are repaired in physiological conditions may not be prevented, leading to extremely deleterious consequences.
The proximity of mtDNA to the OXPHOS system makes mtDNA particularly vulnerable to damage and mutations (oxidized bases and DNA strand breaks) produced by reactive oxygen species (ROS), which are intrinsic OXPHOS byproducts. BER is the primary pathway to repair ROS-induced mtDNA lesions. The mtDNA repair system is less efficient and less known than the nuclear one. Several enzymes take part in the BER-mediated repair process. The latter step, mediated by ligase III, is kinetically the rate-limiting phase in mitochondrial BER. Therefore, cells overexpressing ligase III respond better to oxidative stress. If BER is crippled, DNA repair intermediates can arise, leading to DNA strand breaks, which block DNA replication and transcription, affecting mitochondrial function.9 The presence of mtDNA damaging conditions, such as oxidative stress, could then trigger mitochondrial and cellular demise, and cause the clinical manifestations.
In conclusion, we identified peculiar cases with a combination of pathogenic variants that distinguish between the mitochondrial and nuclear function of ligase III. This genotype allows us to conclude that the mitochondrial dysfunction led to disease in our patients. Moreover, we confirmed that LIG3 pathogenic variants are associated with MDS but also expanded the clinical spectrum from a MNGIE-like syndrome with infantile/teenage onset to neonatal fatal myopathy.
Data availability
Data about the analyses on mitochondrial DNA are available on the repository Zenodo: https://doi.org/10.5281/zenodo.4923605.
For data related to the multi-omics analyses, refer to Kopajtich et al.3
Acknowledgements
The ‘Cell line and DNA Bank of Genetic Movement Disorders and Mitochondrial Diseases’ of the Telethon Network of Genetic Biobanks (grant GTB12001J) and the EurobiobanK Network supplied biological specimens.
Funding
This study was supported by the BMBF (01GM1920A), the Italian MoH (J42F19000030006-RE17) and Horizon 2020 (N°825575) through the European Joint Programme on Rare Diseases (EJP RD) project GENOMIT; by the BMBF (01KU2016A) and the Italian MoH (J49C2000019000-RE15) through the ERA PerMed project PerMiM; BMBF grant to the German Network for Mitochondrial Disorders (mitoNET, 01GM1906D). This project was carried out in the Center for the Study of Mitochondrial Pediatric Diseases funded by the Pierfranco e Luisa Mariani Foundation.
Competing interests
The authors report no competing interests.
References
- 1. Bonora E, Chakrabarty S, Kellaris G, et al. Biallelic variants in LIG3 cause a novel mitochondrial neurogastrointestinal encephalomyopathy. Brain. 2021;144(5):1451–1466. [DOI] [PubMed] [Google Scholar]
- 2. El-Hattab AW, Scaglia F.. SUCLG1-related mitochondrial DNA depletion syndrome, encephalomyopathic form with methylmalonic aciduria. In: Adam MP, Ardinger HH, Pagon RA, eds. GeneReviews®. University of Washington, Seattle; 2017. [PubMed] [Google Scholar]
- 3. Kopajtich R, Smirnov D, Stenton SL, et al. Integration of proteomics with genomics and transcriptomics increases the diagnostic rate of Mendelian disorders. bioRxiv. [Preprint] doi:10.1101/2021.03.09.21253187
- 4. Yépez VA, Gusic M, Kopajtich R, et al. Clinical implementation of RNA sequencing for Mendelian disease diagnostics. bioRxiv. [Preprint] doi:10.1101/2021.04.01.21254633 [DOI] [PMC free article] [PubMed]
- 5. Lakshmipathy U, Campbell C.. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res. 1999;27(4):1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Viscomi C, Zeviani M.. MtDNA-maintenance defects: Syndromes and genes. J Inherit Metab Dis. 2017;40(4):587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Puebla-Osorio N, Lacey DB, Alt FW, Zhu C.. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol Cell Biol. 2006;26(10):3935–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shokolenko IN, Fayzulin RZ, Katyal S, McKinnon PJ, Wilson GL, Alexeyev MF.. Mitochondrial DNA ligase is dispensable for the viability of cultured cells but essential for mtDNA maintenance. J Biol Chem. 2013;288(37):26594–26605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Akbari M, Keijzers G, Maynard S, et al. Overexpression of DNA ligase III in mitochondria protects cells against oxidative stress and improves mitochondrial DNA base excision repair. DNA Repair (Amst.). 2014;16:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Legati A, Zanetti N, Nasca A, et al. Current and new next-generation sequencing approaches to study mitochondrial DNA. J Mol Diagn. 2021;23(6):732–741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data about the analyses on mitochondrial DNA are available on the repository Zenodo: https://doi.org/10.5281/zenodo.4923605.
For data related to the multi-omics analyses, refer to Kopajtich et al.3