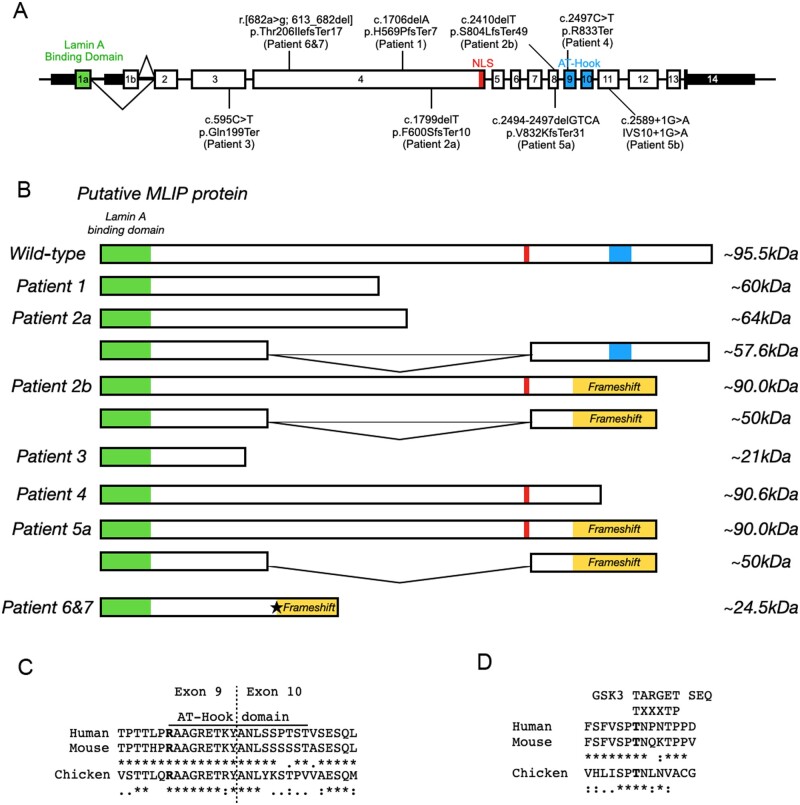
Figure 2.
Localization and effect of MLIP variants. (A) MLIP genomic map with identified position of each pathogenic variant. (B) Expected effects of the MLIP pathogenic variants on the size of MLIP protein for the various patients. Star: Non-canonical 3′ splice site in exon 4. (C and D) Alignment of MLIP between species in the AT-Hook domain (C) and GSK3 phosphorylation motif (D). The consensus sequence for C and D is denoted by: asterisk as identical; a single dot as conserved substitutions; and double dot as semi-conservative substitutions.