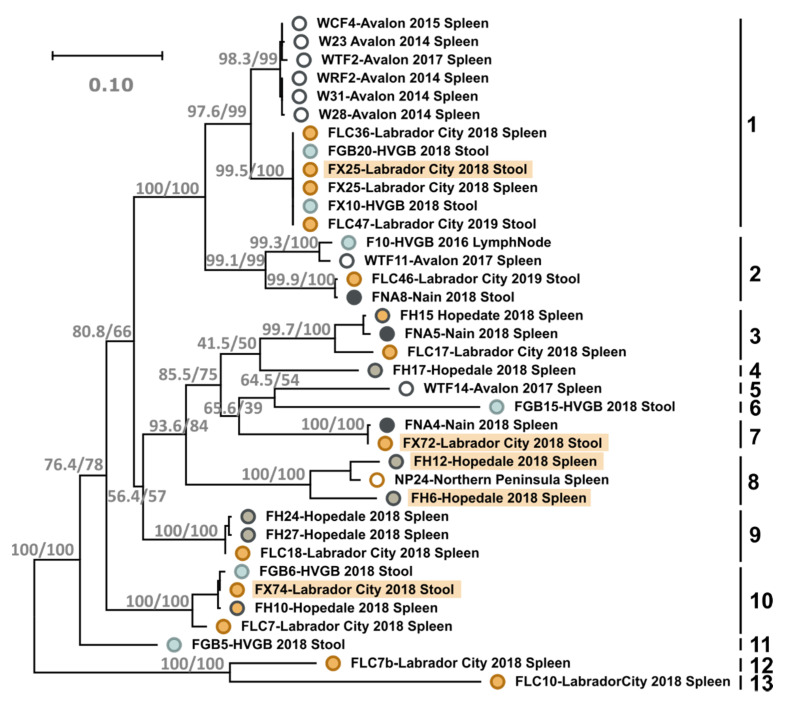
Figure 4.
Phylogenetic analysis of Newlavirus strains identified in this study. The unrooted phylogenetic tree is based on partial VP1 nucleotide sequences and was built with the maximum-likelihood method based on the GTR + F+I + G4 model with IQ-Tree. The outcomes of the SH-aLRT and bootstrap test (1000 replicates) are shown for the main nodes. The viruses identified in this study are labelled with colored circles corresponding to the sampling locations. Each sequence name includes the viral strain identifier, sampling collection site and type of sample in which the virus was identified. The 13 viral genotypes are indicated on the right and strains whose complete genomes were sequenced are highlighted in orange. HVGB: Happy Valley-Goose Bay.