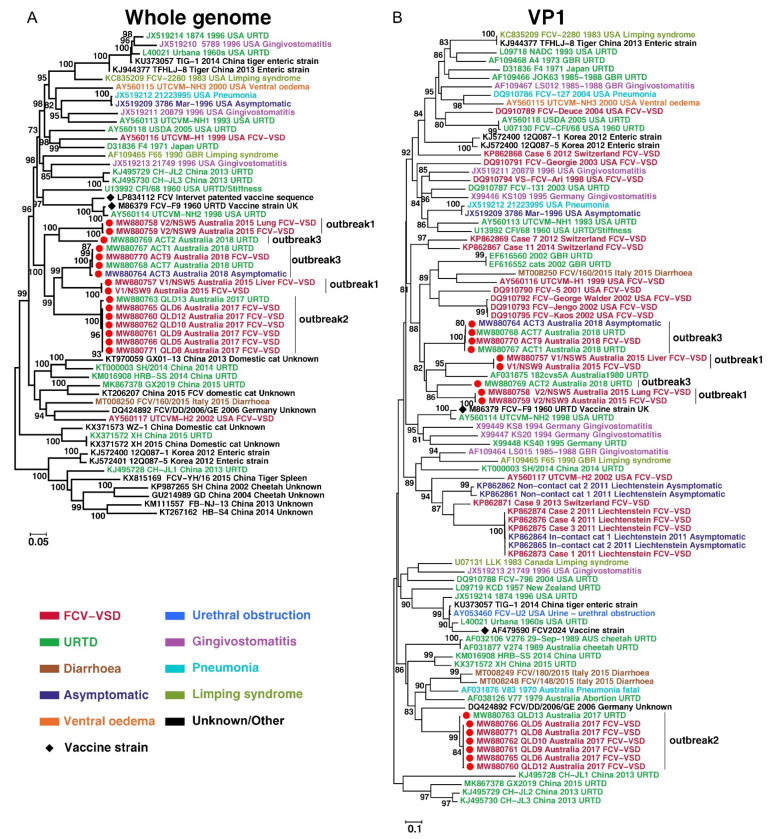
Figure 1.
Evolutionary history of viruses discovered in this study. Maximum-likelihood trees were reconstructed based on (A) Whole-genome and (B) partial VP1 gene alignments, which included virus sequences obtained in this study (marked with solid red circles) as well as those obtained from the GenBank. The trees were mid-point rooted and bootstrap values of ≥70% are marked on the tree. Each sequence name contains the accession number, strain name, geographic location, host and isolation year, if available. Disease phenotype is colour coded.