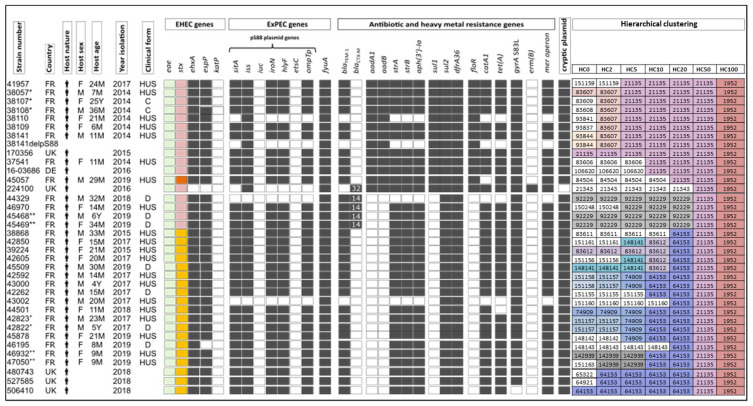
Figure 2.
SNP-Phylogeny and hierarchical clustering of 35 clinical O55:H9 EHEC isolated from Humans in Europe between 2014 and 2019. Caption: The origin of the strain (country, year, and source of isolation) is represented, if available, by the suitable abbreviation (DE, Germany; FR, France; UK, United Kingdom) and the symbol of a human. The clinical form is indicated by a code as follows: HUS, hemolytic uremic syndrome; C, carrier; and D, (bloody)-diarrhea. Demographic and clinical data are missing for the non-French strains. Among these strains, seven are linked, as they were isolated from three families (38057*, 38107*, and 38108*; 45468** and 45469**; and 45822+ and 45823+). Two strains were isolated from the urine and stool from the same patient without signs of urosepsis (46932++ and 47050++, respectively). The presence of intimin (eae) and Shiga toxin (stx) genes is represented by a colored square, depending on their subtype (eae-ξ, green; stx2a, yellow; stx2d, pink; and stx2f, orange). The presence of virulence and antibiotic-resistance genes or the cryptic plasmid (pR444_B) is indicated by a black square. The numbers in the squares indicate the variant of blaCTX-M. The genomes were also assigned to hierarchical clusters (HCs) based on core genome MLST to assess their genomic relatedness. Common HCs are represented by a number highlighted in the same color, indicating that the strains are closely related. For example, two strains sharing the same HC10 means they have links that are no more than 10 alleles apart. 38141delpS88 is the 38,141 mutant cured of its pS88-like plasmid using SDS.