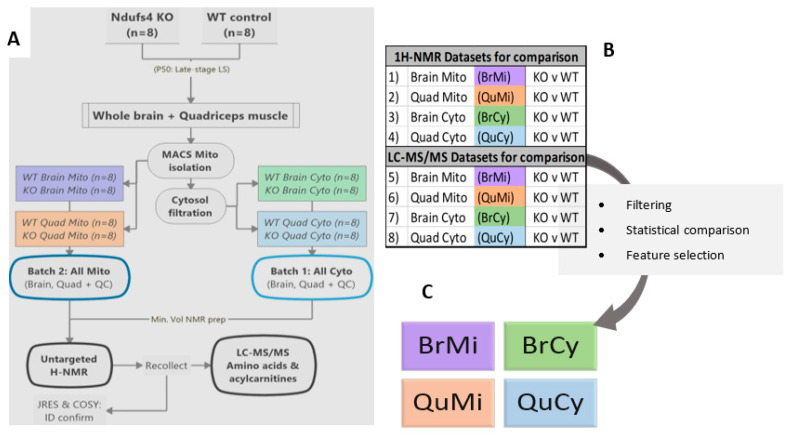
Figure 1.
Summarized workflow for the study. Ndufs4 and control mouse quadriceps and whole brains were subjected to MACS mitochondrial isolation and cytosolic filtering, before subsequent 1H-NMR and LC-MS/MS analyses (A). Datasets were extracted as such that Ndufs4 KO and WT were compared in each matrix (B), before data processing and statistical feature selection. Thereafter, the combined significant feature (VIP) datasets for each tissue and compartment were further processed and interpreted (C).