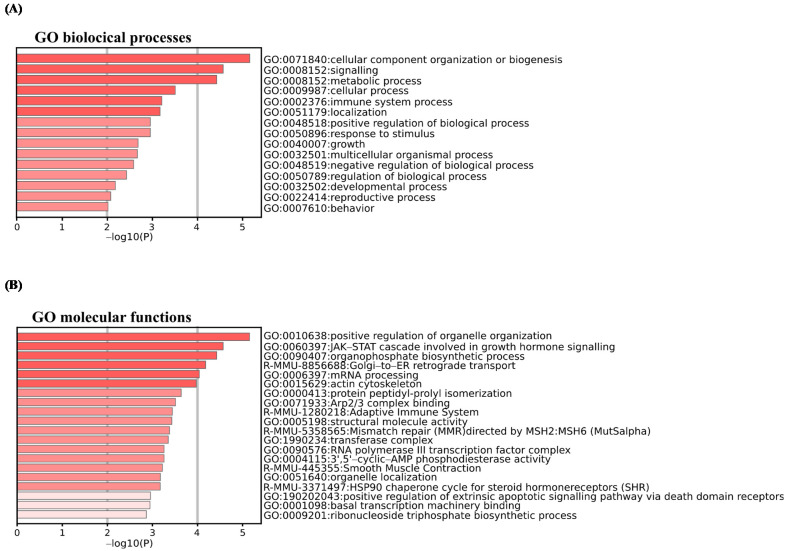
Figure 3.
Pathway and process enrichment analysis using Metascape database for DEPs. Enriched pathways and processes were identified using Metascape database (http://metascape.org, (accessed on 7 July 2021)) for all DEPs from ANOVA test and with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Gene Ontology (GO) biological processes, GO cellular component, GO molecular function, Reactome Gene Sets, and CORUM as ontology sources. Top enrichment biological processes (A) and molecular functions (B) term clusters, colored by −log10 (p-value). Threshold: 0.3 kappa score; similarity score > 0.3. The darker the color is, the more significant the term. Significant terms were then hierarchically clustered into a tree based on kappa—statistical similarities among membership similarities. Then, the 0.3 kappa score was applied as the threshold to cast the tree into term clusters. More specifically, the most statistically significant term within a cluster was selected for cluster annotation. The first 20 significant most representative term clusters are shown.