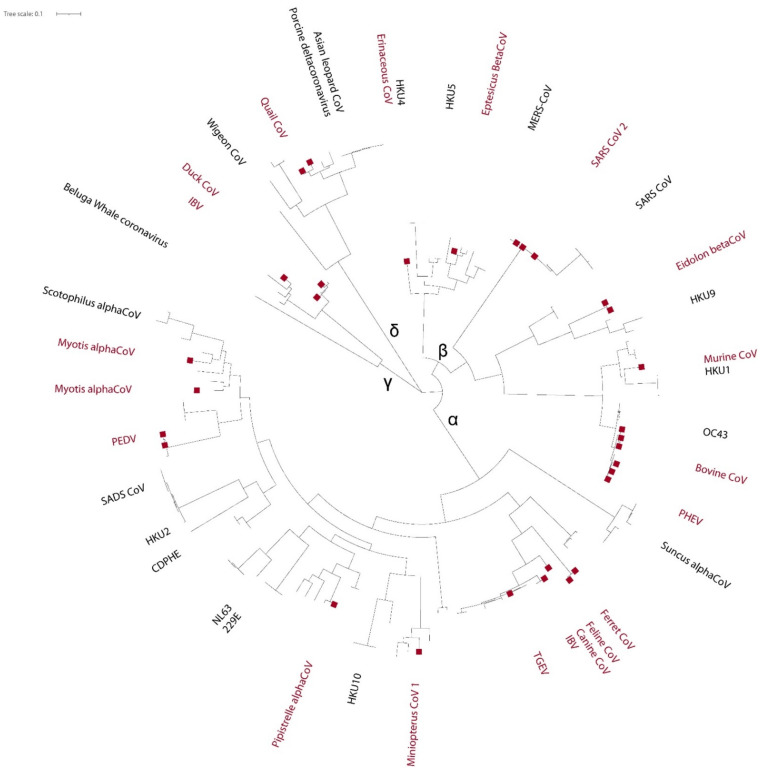
Figure 4.
ML phylogenetic tree of coronaviruses. Sequences were aligned using the G-INS1 and default parameters implemented in Mafft using online tool: https://mafft.cbrc.jp/alignment/server [50]. ML phylogenetic trees were inferred using PhyML (version 3.0) implemented in Seaview (Lyon, France), employing the GTR+G4 substitution model, a heuristic SPR branch-swapping algorithm, and SH-like branch supports [51]; obtained trees were edited online for graphical display using iTOL on server: https://itol.embl.de/ [52]. CoV species investigated in the study are indicated in red, with branches of tested strains shown as red boxes.