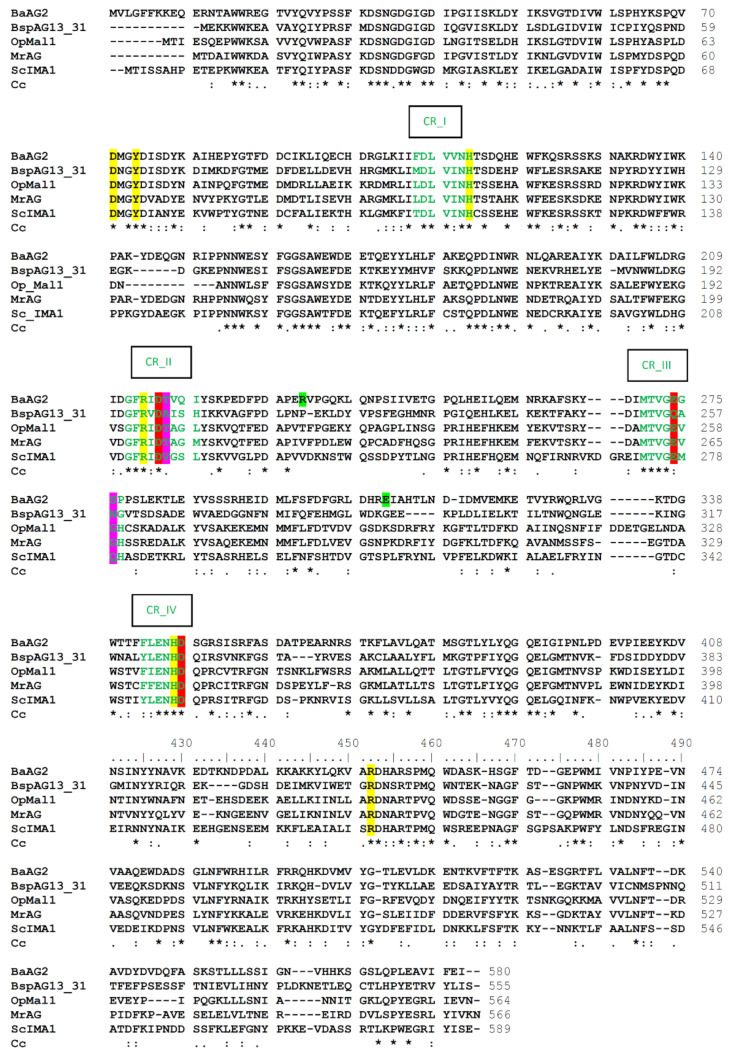
Figure 2.
Clustal Omega alignment of amino acid sequences of BaAG2 (GenBank: MZ467078, translation), BspAG13_31 (α-glucosidase of Bacillus sp. AHU2216; PDB: 5ZCE), OpMal1 (maltase-isomaltase Mal1 of O. polymorpha; GenBank: XP_018213389.1, AWO14629.1), MrAG (α-glucosidase of M. reukaufii; GenBank: QLP89119.1) and ScIMA1 (isomaltase of S. cerevisiae; GenBank: NP_011803.3). Catalytic triad residues are shown with a red background, residues corresponding to BaAG2 residues making contacts with the substrate at −1 subsite (see also Figure 4) with a yellow background, further binding sites residues of BaAG2 with a green background, and residues equivalent to Gln279 and Val216 of ScIMA1 determining substrate specificity have a magenta background. Four conserved regions (CR) of α-glucosidases as given by [49] on the basis of Taka-amylase are shown by green letters. Clustal consensus (Cc) is marked below the sequence indicating conservation — * positions with fully conserved residue; : positions with residues of strongly similar properties; . positions with residues of weakly similar properties.