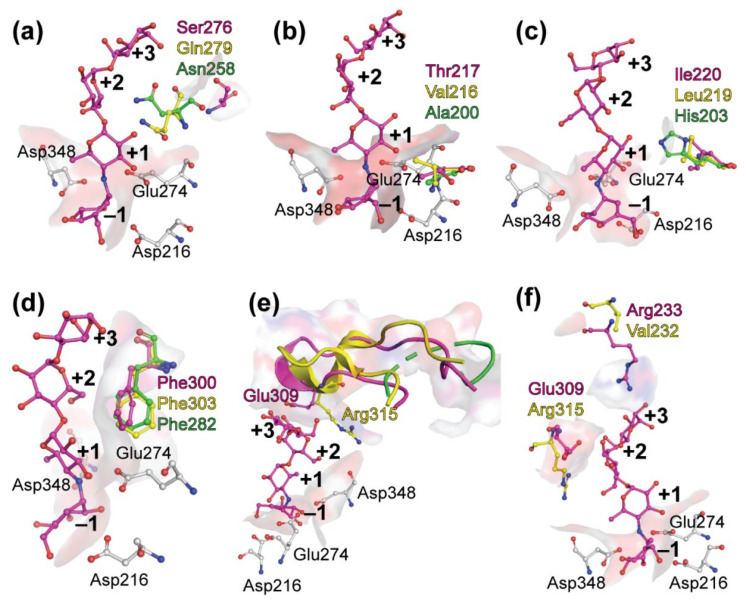
Figure 5.
Amino acids participating in substrate binding among GH13 α-glucosidases. Positions of Ser276, Thr217, Ile220, Phe330, Glu309 and Arg233 of BaAG2 from B. adeninivorans (in magenta) shown on panels a–f, respectively, were superimposed with the Glu277Ala mutant of S. cerevisiae isomaltase IMA1 (in yellow; PDB: 3AXH) and the Glu256Gln mutant of Bacillus sp. AHU2216 maltase BspAG13_31 (in green, PDB: 5ZCE). The catalytic triad (Asp216, Glu274 and Asp348) of BaAG2 is shown by sticks in grey, and acarbose from the BaAG2 structure is colored in magenta. The surface mode is applied to amino acids from BaAG2-acarbose structure and subsites of catalytic cavity, from −1 to +3, are designated with bold numbers. Structures were aligned and visualized with PyMol [39].