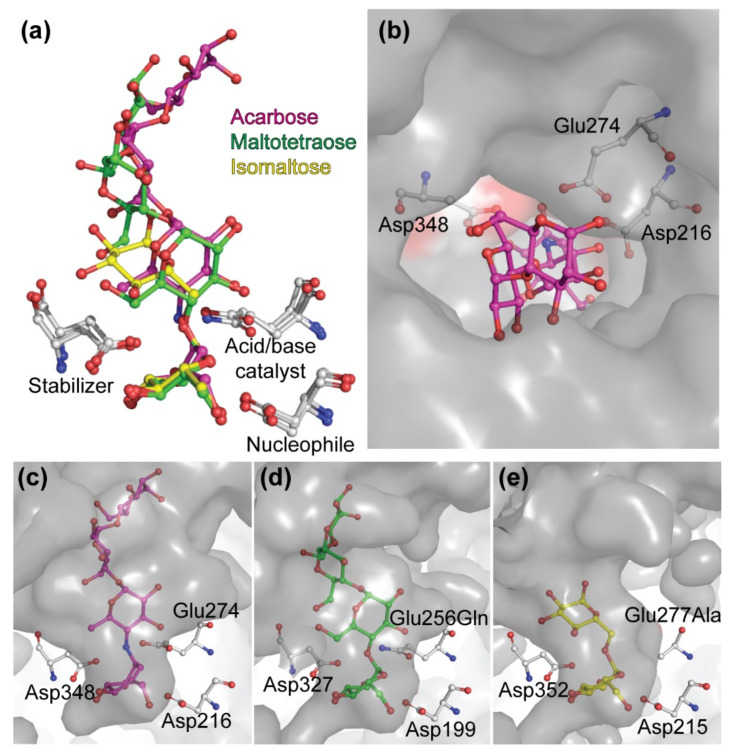
Figure 6.
Substrate binding pocket of selected GH13 enzymes. Superimposed conformations of acarbose, maltotetraose and isomaltose (a). Top view of substrate binding cavity of the maltase BaAG2 of B. adeninivorans with bound acarbose (magenta) (b). Side views of substrate binding modes of acarbose (panel c, magenta) to BaAG2, maltotetraose (panel d, green) to BspAG13_31 mutant Glu256Gln of Bacillus sp. AHU2216 (PDB: 5ZCE) and isomaltose (panel e, yellow) to IMA1 mutant Glu277Ala of S. cerevisiae (PDB: 3AXH). Catalytic triad amino acids are marked by grey sticks. Structures were aligned and visualized with PyMol [39].