Abstract
We determined the kinetics of anti–SARS-CoV-2 antibody response in fifteen hospitalized COVID-19 patients. Patients were divided into mild/moderate (mild, n = 1; moderate, n = 4) or severe (n = 10) and virus-specific anti–Nucleocapsid IgM, anti–Spike IgG and anti–Spike IgA were measured in serial serum samples collected 0 to 15 days after hospital admission. Surrogate neutralization assays were performed by testing inhibition of ACE-2 binding to Spike. In 3 patients (2 severe and 1 moderate case), serum antibodies and T-cell memory were monitored 6 months after baseline. Although IgM response tended to appear first, patients affected by less severe disease were more prone to an early IgG/IgA response. Neutralization of Spike binding to ACE2 correlated with anti–Spike IgG and IgA. IgG and IgA antibody response persisted at the 6 months follow-up. A recall T-cell response to the Spike antigen was observed in 2 out of 3 patients, not related to disease severity.
Keywords: SARS-CoV-2, COVID-19, Serology, Disease Severity, T-cell immunity
1. Introduction
The immune response against SARS-CoV-2 remains unclear and many caveats in its knowledge are still present. A correlation of clinical severity with high titres of SARS-CoV-2 antibodies, mainly immunoglobulin (Ig)G, was shown (Zhao et al, 2020; Long et al, 2020; Ma et al, 2020), and the importance of neutralizing humoral immunity on disease progression has been shown (Garcia-Beltran et al., 2021). Fewer studies assessed serum SARS-CoV-2 IgA; this Ig subclass becomes detectable 4 to 6 days after symptom onset with a higher and more persistent serum level than IgM (Padoan et al, 2020; Yu et al, 2020). IgA levels seem significantly higher in severe than in mild/moderate SARS-CoV-2 infections (Yu et al, 2020; Bartsch et al., 2021).
Early antibody responses appear to be correlated with disease outcome (Atyeo et al., 2020). Timing of IgM and IgG seroconversion is controversial, with serum IgM appearing before IgG or synchronously (Zhao et al, 2019; Xiang et al, 2020), or even later (To et al., 2020, Zhang et al., 2020). There is growing evidence of pre-existing humoral and T-cell immunity caused by exposure to common cold coronaviruses (Ng et al., 2020; Sette and Crotty, 2020). Immunity to SARS-CoV-2 is also related to T-cell responses (McMahan et al, 2021). Patients who resolve infection have been shown to generate virus‐specific T‐cell responses (Sekine et al, 2020; Weiskopf et al, 2020). It has been proposed that SARS-CoV-2 induces multifunctional memory T cells which play a role in preventing recurrent episodes of severe SARS-CoV-2 infections (Sekine et al, 2020; Grifoni et al, 2020).
The objective of the study was the characterization of the serologic response to SARS-CoV-2 in patients hospitalized with confirmed SARS-CoV-2 infection. We aimed at comparing the kinetics of IgM, IgG, IgA, and virus neutralization activity in a group of patients affected by mild/moderate or severe SARS-CoV-2 infection. Six months after baseline, SARS-CoV-2 specific antibodies, and T-cell responses were measured in a smaller number of individuals.
2. Material and methods
2.1. Clinical sample
Fifteen patients hospitalized with confirmed COVID-19 at the Infectious Diseases division of Policlinico Umberto I Hospital, Rome, Italy, were included in the study (Table 1 ). The study was approved by local Ethic Committee and all participants have signed an informed consent. At the hospital admission all patents were affected by COVID-19 related pneumonia and, according to the PaO2/FiO2 ratio (partial oxygen pressure/oxygen flow), 10 patients were classified as having a severe disease (PaO2/FiO2 < 250), 4 having a moderate disease (PaO2/FiO2 > 250) in need of oxygen supply, and 1 having mild disease(PaO2/FiO2 > 250, no need of oxygen supply). Patients were re-tested for SARS-CoV-2 RNA 6 days after hospital admission and then every 2 days, until a negative nasopharingeal (NP) swab result was obtained.
Table 1.
Demographic and clinical characteristics of enrolled subjects.
| Patient Nr. | Sex | Age, years | Comorbidities | Days from symptoms onset to hospital admission | Clinical severity | Oxygen supply during hospital stay | CT value E gene | CT value S gene | Days to SARS-CoV-2 negative NP swab | Length of hospital stay, days |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 64 | AI, DM | 10 | Mild | None | 32.9 | 32.2 | 9 | 19 |
| 2 | M | 48 | NHL | 2 | Moderate | VM | 29.8 | 28.9 | 14 | 16 |
| 3 | M | 83 | AI, IHD | 7 | Moderate | VM | 28.2 | 27.1 | 17 | 35 |
| 4 | M | 42 | DM | 6 | Moderate | VM | 32.6 | 34.2 | 13 | 19 |
| 5 | M | 58 | AI | 13 | Moderate | VM | 32.1 | 31.0 | 12 | 19 |
| 6 | M | 62 | None | 6 | Severe | HFNC/CPAP | 22.2 | 21.2 | NA | 27 |
| 7 | M | 78 | Lung cancer | 1 | Severe | HFNC/CPAP | 14.9 | 14.8 | NA | 28 |
| 8 | M | 64 | None | 10 | Severe | HFNC/CPAP | 23.8 | 23.2 | 25 | 28 |
| 9 | F | 60 | AI | 4 | Severe | HFNC/CPAP | 20.3 | 19.3 | 17 | 21 |
| 10 | M | 69 | None | 3 | Severe | HFNC/CPAP | 31.0 | 32.0 | 12 | 14 |
| 11 | M | 52 | None | 3 | Severe | HFNC/CPAP | 23.6 | 22.9 | 25 | 30 |
| 12 | M | 78 | AI | 8 | Severe | HFNC/CPAP | 35.7 | 32.2 | 15 | 47 |
| 13 | M | 66 | None | 5 | Severe | HFNC/CPAP | 27.4 | 28.1 | 11 | 21 |
| 14 | M | 76 | AI, COPD, HT | 12 | Severe | HFNC/CPAP | 26.9 | 26.1 | 25 | 38 |
| 15 | F | 63 | None | 7 | Severe | HFNC/CPAP | 31.0 | 30.2 | 12 | 19 |
AI = arterial hypertension; NHL = non–hodgkin Lymphoma; COPD = chronic obstructive pulmonary disease; IHD = ischemic heart disease; DM = diabetes mellitus; HT = hypothyroidism; HFNC = high flux nasal cannula; CPAP = continuous positive airway pressure; VM = venturi mask; NA = not-applicable (patients died).
2.2. Serology
Serum samples were available at 3 different time-points: at hospital admission (T0), 5 to 10 days (T1), and 11 to 15 days (T2) after hospital admission. Immediately after collection, sera were shipped to the Department of Infectious Diseases, Istituto Superiore di Sanità (ISS); 0.5 mL aliquots were prepared and stored at -80°C. Serology testing was performed at ISS by personnel blinded to specimen classification within 1 month from serum collection.
ELISA assays were used to measure Anti–Nucleocapsid IgM (Dia-Pro diagnostics Bioprobes, Sesto San Giovanni (MI), Italy), anti–Spike IgG and anti–Spike IgA (both from Euroimmun, Lübeck, Germany), according to manufacturers’ instructions. A surrogate neutralization assay (GenScript, Leiden, The Netherlands) was used to detect the virus neutralizing ability of patients’ sera. The assay detects and measures neutralizing antibodies blocking the Spike RBD-ACE2 interaction in vitro and was used according to manufacturer's instructions.
2.3. T-cell response
Additional blood samples from patients #4, #11 and #15 were collected 6 months after hospital discharge and humoral and T-cell immune responses to SARS-CoV-2 assessed. Since the second wave of Covid-19 cases hit the Policlinico Umbero I hospital at the end of September 2020, it was not possible to include in the 6-month follow-up study other patients, due to safety concerns. Virus-specific T-cell responses were measured by stimulating patients’ peripheral blood mononuclear cells (PBMCs) with overlapping peptides covering the immunodominant sequence domains of the Spike protein (Miltenyi, Bergisch Gladbach, Germany). After overnight stimulation, cells were incubated with Live/Dead fixable violet dead cell stain kit used to exclude dead cells from the analyses (Thermo Fisher Scientific, Waltham, MA). Cells were then fixed in paraformaldehyde 2%, permeabilized using a solution containing saponine 0.5% and stained with a predetermined optimal concentration of fluorochrome-conjugated Abs: anti–CD3-Horizon-BV510, anti–IL-2-FITC, anti–TNFα PE-Cy7 (all from BD Biosciences, Franklin Lakes, NJ), anti–IL-4-APC, anti–IFN-γ-PerCP-Cy5.5 (both from Biolegend, San Diego, CA), anti–CD8-PE (eBiosciences, Thermo Fisher Scientific). Cells were then acquired by flow cytometry in a Gallios Flow Cytometer (Beckman Coulter, Indianapolis, IN) and compensation matrices were calculated with Kaluza Analysis software (Beckman Coulter).
3. Results
3.1. Clinical sample
Fifteen patients were included in the study (Table 1). Patients with mild/moderate SARS-CoV-2 infection had a lower mean age (59.0 vs 66.8; P = 0.27) and higher median Ct values of PCR from NP samples (gene E: 32.10 vs 25.35, P = 0.05; gene S: 31.00 vs 24.65, P = 0.058) than those with severe infection. As expected, severely ill patients required more days for a negative result of NP swab (median 16.0 vs 13.0; P = 0.29).
3.2. Anti–SARS-CoV-2 IgM, IgG, IgA antibody responses
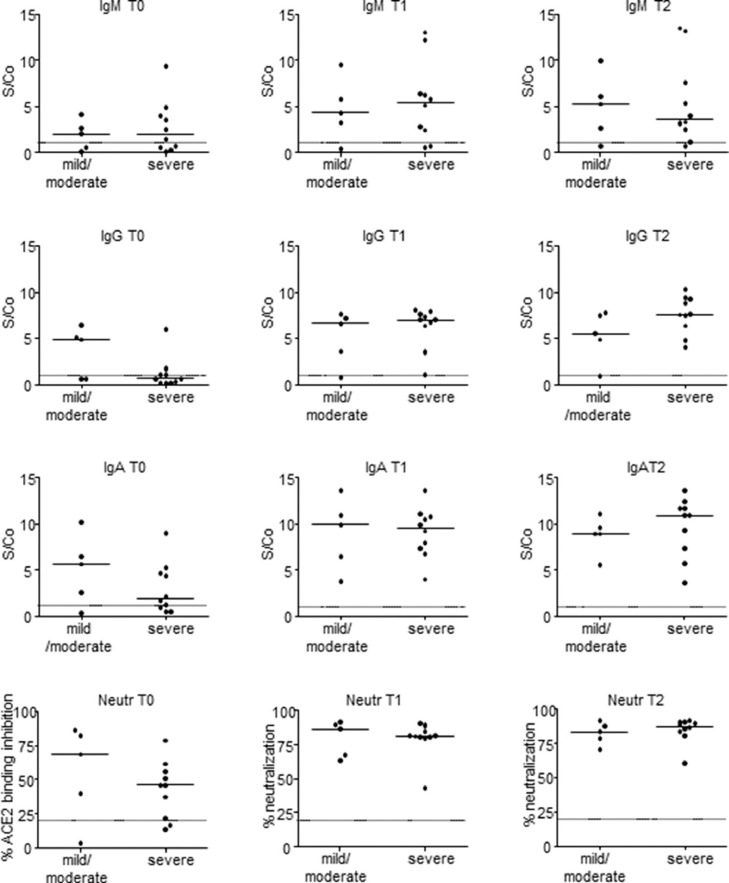
Serum samples were collected at hospital admission (T0), 5 to 10 days (T1), and 11 to 15 days (T2) after hospital admission. At T0, IgM specific for the viral nucleocapsid protein were above the positivity threshold in 9 patients (60.0 %). At T1, a positive IgM response was measured in 12 patients (80.0 %); at T2, 13 patients (86.7%) had a positive IgM response. The IgM response showed a slightly higher median value among severely affected patients at T1 and lower at T2 (Fig. 1 ).
Fig. 1.
SARS-CoV-2 specific antibody response in mild/moderate and severe COVID-19 patients. Serologic anti–Nucleocapsid IgM, anti–Spike IgG, and anti–Spike IgA antibodies were tested by enzyme-linked immunosorbent assay. Sera neutralization activity was assessed by surrogate virus neutralization test based on antibody-mediated blockage of ACE2-Spike protein-protein interaction. Solid horizontal lines represent Mean. Dotted lines represent positivity thresholds. T0: hospital admission; T1: 5 to 10 days after hospital admission; T2: 11 to 15 days after hospital admission; S/Co: Signal to Cut-off (S/Co) ratio.
Anti–Spike IgG at T0 were above the positivity threshold in 5 patients (13.3%). At T1 a positive IgG response was measured in 13 patients (86.7%); at T2 14 patients (93.3%) had a positive IgG response. When patients were classified according to disease severity, we found that at T0 the median IgG response was higher in mild/moderate patients, although this difference was not statistically significant. IgG levels were higher among severely affected patients at T2 (Fig. 1).
Anti–Spike IgA at T0 were above the positivity threshold in 11 out of 15 patients (73.3%). At T1 and T2 a positive IgA response was measured in all the patients. Higher IgA levels were found at T0 in patients with a mild/moderate disease (Fig. 1).
3.3. SARS‐CoV‐2 neutralization correlates with IgG and IgA serum levels in laboratory‐confirmed COVID‐19 patients
At hospital admission Spike RBD-ACE2 neutralization activity was measured in 13 patients (86.7%). At T1 and T2 a neutralizing response was measured in all the patients. Patients with mild/moderate disease presented higher neutralizing antibody levels at hospital admission as compared to severe patients, although this was not statistically significant (Figure 1).
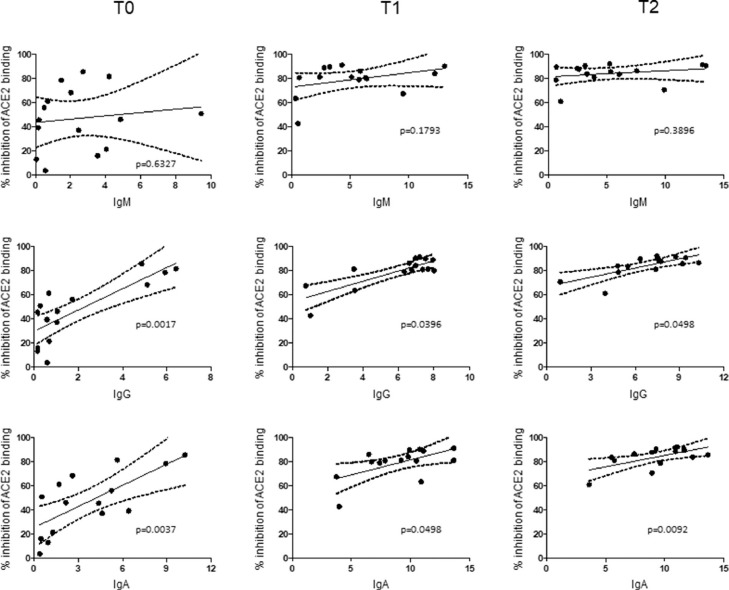
A statistically significant positive correlation of in vitro neutralization of Spike RBD-ACE2 binding with IgG and IgA serum levels at all the time-points was found, whereas no correlation was evident between anti–Nucleocapsid IgM and Spike RBD-ACE2 neutralizing activity (Fig. 2 ).
Fig. 2.
Serum IgG and IgA levels correlate with inhibition of SARS-CoV-2-ACE2 binding. Linear regression correlating serum levels of anti–Nucleocapsid IgM (top), anti–Spike IgG (middle) and, anti–Spike IgA (bottom) antibodies with Spike-ACE2 binding neutralization; numerical values along horizontal axis represents Signal to Cut-off ratios for each serologic assay. Dotted lines represent 95% confidence band. P values are shown.
3.4. Persistence of anti–SARS-CoV2 immunity 6 months after the infection
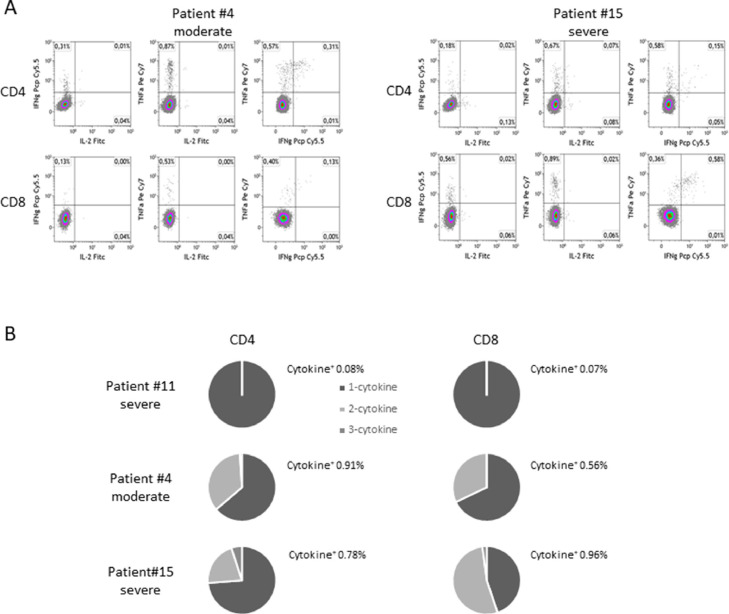
A 6-month follow-up analysis was performed on 3 out of 15 patients. We did not observe any decline in anti–Spike IgG levels (T2 mean ± SD: 6.53 ± 0.27; 6-month mean ± SD: 6.21 ± 0.9). Anti–Spike IgA levels declined but were still above the positivity threshold (T2 mean ± SD: 11.70 ± 0.56; 6-month mean ± SD: 4.13 ± 2.16). Sera retained surrogate neutralization activity (data not shown). Two patients (1 moderate and 1 severe case) were able to mount a virus-specific T-cell response, marked by the production of TNF-α and IFN-γ by CD4 and CD8 T cells (Fig. 3 ), while 1 patient was unresponsive. Profiles of T-cell response were rather different. A higher frequency of cytokine positive CD4+ versus CD8+ T cells (0.91% vs 0.56%) was found in patient #10 (moderate), while in patient #15 (severe) this ratio was inverted (0.78% vs 0.96%). The proportion of CD4 and CD8 cells producing 1, 2 or 3 cytokines was also different in the 2 responsive patients.
Fig. 3.
SARS-CoV-2 specific T-cell response 6 months after recovery. (A) Dot plots show the percentage of CD4+ and CD8+ T cells producing IFN-γ, TNF-α and IL-2 in response to a pool of overlapping peptides covering the immunodominant sequence domains of Spike protein. (B) Frequencies of mono- and multifunctional specific CD4+ and CD8+ T-cell responses.
4. Discussion
The correlation of antibody responses to SARS-CoV-2 with COVID-19 disease severity is still unclear. Our data suggest that individuals with an earlier appearance of virus-specific IgG and IgA potentially have a better outcome. In patients with a mild/moderate disease, IgG responses tended to occur simultaneously with IgM or even slightly earlier, while in patients with severe disease IgG and IgA were produced later during the course of the disease. Both IgG and IgA levels were correlated to serum neutralizing activity.
Our observations agree with a recent study by Carsetti and co-workers (2020) who found that IgA and IgG produced relatively late in the course of the infection characterize severe disease (Carsetti et al, 2020). Moreover, it has been shown that patients with more severe illness display higher antibody titers than those with milder disease (Long et al, 2020; Röltgen et al, 2020).
The presence of anti–SARS-CoV-2 IgG and IgA at T0 in mild/moderate patients suggests that some pre-existing immunity to the 4 endemic coronaviruses causing common cold in humans (HCoV-229E, -NL63, -OC43, and -HKU1) may play a role in counteracting the infection. These findings are supported by a recent study showing that COVID-19 disease appears less severe in patients with endemic coronavirus infection (Sagar et al, 2021).
5. Limitations
Before drawing conclusions, limitations of the study should be mentioned. The number of patients enrolled is small and probably not sufficient for making strong conclusions. Only 3 patients were used in the cellular immunity studies at 6 months. Although incomplete and preliminary, the 6-month follow-up showed the persistence of serologic response in 3 patients. Data on T-cell response suggested the possibility of differential responses that could reflect asymptomatic exposure.
6. Conclusion
In conclusion, our data confirm the notion that an early IgG/IgA response focused on the Spike protein may help to counteract the viral infection, leading to a milder course of the disease. Further studies are required to investigate more in depth the duration and patterns of T-cell memory responses.
Acknowledgments
The authors wish to thank the ISS working group on COVID-19: Stefano Fiore, Concetta Fabiani, Eleonora Benedetti, Angela Di Martino, Laura Calzoletti, Marzia Facchini, Giuseppina Di Mario, Giulietta Venturi, Claudia Fortuna, Antonello Amendola, Giulia Marsili.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors have nothing to disclose.
Authors’ contributions
Giorgio Fedele: conceptualization, data curation, writing-original draft preparation and revised version; Gianluca Russo: conceptualization, data curation, writing-original draft preparation and revised version; Ilaria Schiavoni: investigation and Writing-Reviewing; Pasqualina Leone: investigation; Eleonora Olivetta: investigation; Valentina Perri, Maria Antonella Zingaropoli, Maria Rosa Ciardi and Patrizia Pasculli: resources; Claudio Maria Mastroianni: supervision; Paola Stefanelli: conceptualization and supervision.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.diagmicrobio.2021.115586.
Appendix. Supplementary materials
References
- Atyeo C, Fischinger S, Zohar T, Slein MD, Burke J, Loos C, et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020 15;53(3):524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch YC, Wang C, Zohar T, Fischinger S, Atyeo C, Burke JS, et al. Humoral signatures of protective and pathological SARS-CoV-2 infection in children. Nat Med. 2021 doi: 10.1038/s41591-021-01263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsetti R, Zaffina S, Piano Mortari E, Terreri S, Corrente F, Capponi C, et al. Different innate and adaptive immune responses to sars-cov-2 infection of asymptomatic, mild, and severe cases. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.610300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran WF, Lam EC, Astudillo MG, Yang D, Miller TE, Feldman J, et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184(2):476–488.e11. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Rydyznski Moderbacher C, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long QX, Tang XJ, Shi QL, Li Q, Deng HJ, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Ma H, Zeng W, He H, Zhao D, Jiang D, Zhou P, et al. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol. 2020;17(7):773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590(7847):630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KW, Faulkner N, Cornish GH, Rosa A, Harvey R, Hussain S, et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370(6522):1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padoan A, Sciacovelli L, Bassl D, Negrini D, Zuin S, Cosma C, et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: a longitudinal study. Clin Chim Acta. 2020;507:164–166. doi: 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K, Powell AE, Wirz OF, Stevens BA, Hogan CA, Najeeb J, et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol. 2020;5(54):eabe0240. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar M, Reifler K, Rossi M, Miller NS, Sinha P, White LF, et al. Recent endemic coronavirus infection is associated with less-severe COVID-19. J Clin Invest. 2021;131(1) doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T, Perez-Potti A, Rivera-Ballesteros O, Stralin K, Gorin JB, Olsson A, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Crotty S. Pre-existing immunity to SARS-CoV-2: the knowns and unknowns. Nat Rev Immunol. 2020;20(8):457–458. doi: 10.1038/s41577-020-0389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KKW, Tsang OTY, Leung WS, Tam AR, Wu TC, Lung DC, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D, Schmitz KS, Raadsen MP, Grifoni A, Okba NMA, Endeman H, et al. Phenotype of SARS-CoV-2-specific T-cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol. 2020;5(48):eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, et al. Antibody detection and dynamic characteritics in patients with COVID-19. Clin Infect Dis. 2020;71(8):1930–1934. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HQ, Sun BQ, Fang ZF, Zhao JC, Liu XY, Li YM, et al. Distinct features of SARS-CoV-2-specific IgA response in COVID-19 patients. Eur Respir J. 2020;56(2) doi: 10.1183/13993003.01526-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody response to SARS-CoV-2 in patients of novel coronavirus diseases 2019. Clin Infect Dis. 2020;71(16):2027–2034. doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.