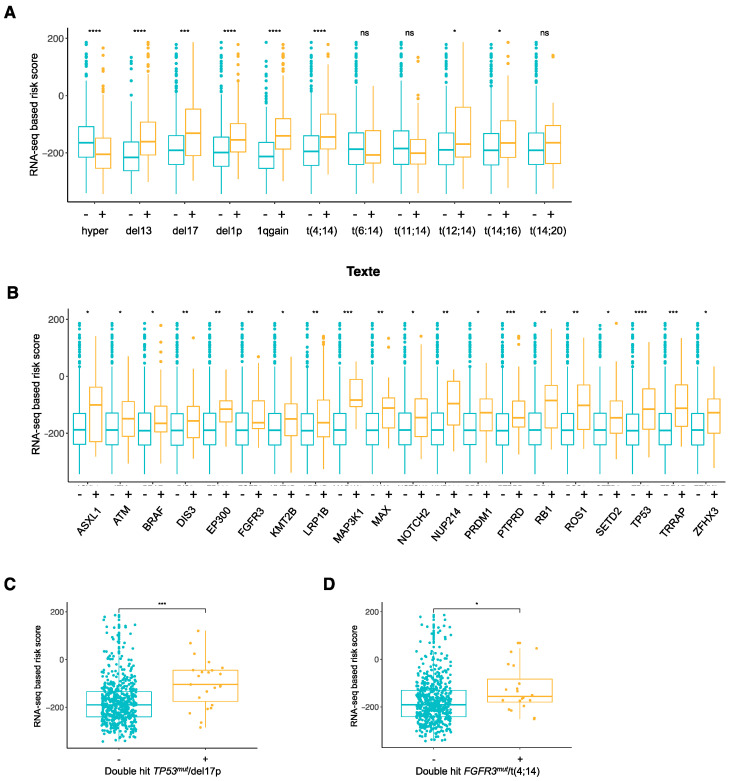
Figure 3.
Association between RNA-seq-based risk scores and cytogenetic abnormalitie subgroups and mutations in MM patients from the CoMMpass cohort. (A) RNA-seq-based risk score in cytogenetic abnormality subgroups. Cytogenetic abnormality subgroups were composed of hyperdyploid (without: n = 240, with: n = 331), del(13p) (without: n = 307, with: n = 266), del(17p) (without: n = 523, with: n = 50), del(1p) (without: n = 455, with: n = 118), 1q gain (without: n = 378, with: n = 195), groups of patients and patients harboring t(4;14) (without: n = 515, with: n = 73), t(6;14) (without: n = 572, with: n = 16), t(11;14) (without: n = 461, with: n = 127), t(12;14) (without: n = 564, with: n = 24), t(14;16) (without: n = 532, with: n = 56), and t(14;20) (without: n = 543, with: n = 45) translocations. (B) Comparison of RNA-seq-based risk scores according to the status of recurrent mutated genes in MM: ASXL1 (−: n = 616, +: n = 15), ATM (−: n = 586, +: n = 45), BRAF (−: n = 576, +: n = 55), DIS3 (−: n = 554, +: n = 77), EP300 (−: n = 614, +: n = 17), FGFR3 (−: n = 592, +: n = 39), KMT2B (−: n = 555, +: n = 76), LRP1B (−: n = 532, +: n = 99), MAP3K1 (−: n = 620, +: n = 11), MAX (−: n = 599, +: n = 32), NOTCH2 (−: n = 596, +: n = 35), NUP214 (−: n = 617, +: n = 14), PRDM1 (−: n = 604, +: n = 27), PTPRD (−: n = 572, +: n = 59), RB1 (−: n = 613, +: n = 18), ROS1 (−: n = 605, +: n = 26), SETD2 (−: n = 597, +: n = 34), TP53 (−: n = 579, +: n = 52), TRRAP (−: n = 605, +: n = 26), and ZFHX3 (−: n = 592, +: n = 39). (C) Comparison of the RNA-seq-based risk score values in MM patients with (n = 23) and without (n = 546) double-hit TP53mut/del(17p). (D) Comparison of the RNA-seq-based risk score values in MM patients with (n = 20) and without (n = 562) double-hit FGFR3mut/t(4;14). Wilcoxon test. ns: nonsignificant, * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001.