Figure 5.
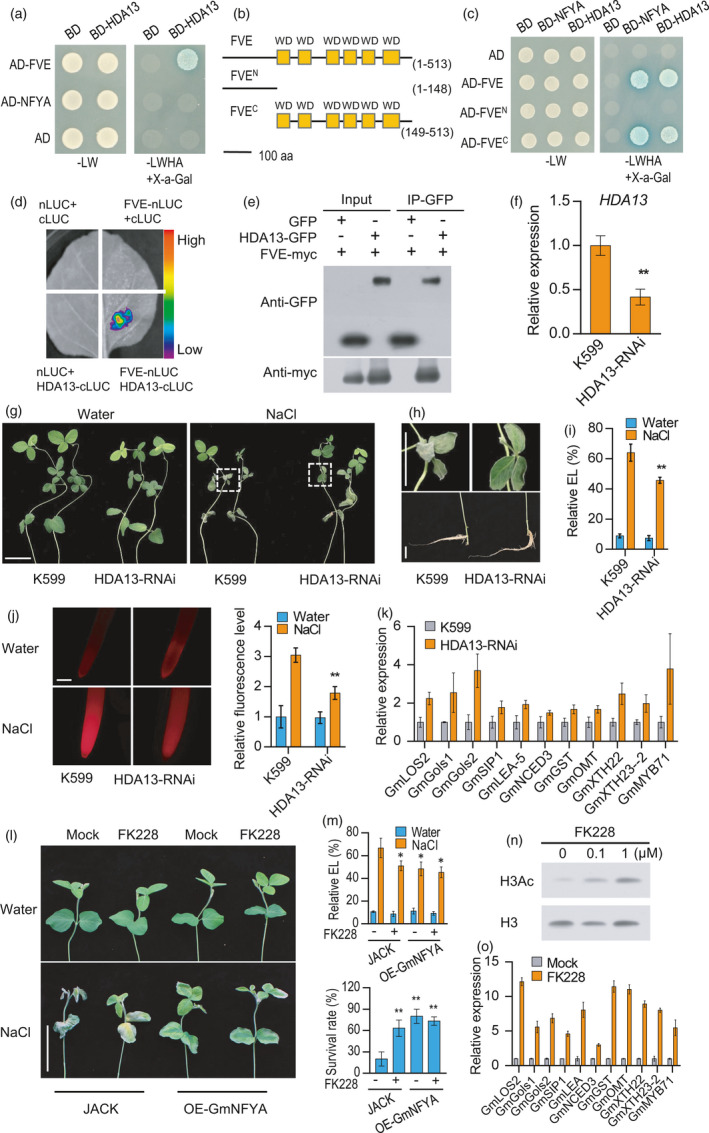
GmHDA13 associates with GmFVE to negatively regulate salt tolerance in soybean. (a) GmFVE interacts with GmHDA13 in a yeast two‐hybrid assay. (b) Different versions of GmFVE used for interaction analysis. Numbers indicate residue positions. WD, WD40 domain. aa, amino acids. (c) GmNFYA and GmHDA13 interact with the C terminal region of GmFVE in yeast cells. (d) Split‐luciferase complementation assay of GmFVE and GmHDA13 interaction in N. benthamiana leaf. (e) Co‐IP assay of the interaction between GmFVE and GmHDA13 in Arabidopsis protoplasts. (f) Expression of GmHDA13 in GmHDA13‐RNAi transgenic hairy roots. HDA13‐RNAi indicates transgenic hairy roots with reduced transcript level of GmHDA13. Bars indicate SD (n = 3). (g) Phenotype of the soybean plants with GmHDA13‐RNAi transgenic hairy roots after salt treatment. Scale bar = 5 cm. (h)The growth of leaf and root after salt treatment from plants in G. Bar = 2 cm. (i) Relative electrolyte leakage of trifoliolate leaves from the plants with GmHDA13‐RNAi transgenic hairy roots after 100 mM NaCl treatment for 3 d. Bars indicate SD (n = 4). (j) ROS levels of the GmHDA13‐RNAi transgenic and control roots after 80 mM NaCl treatment for 6h. Scale bar = 50 μm. (k) Expression of GmNFYA‐regulated genes in GmHDA13‐RNAi transgenic hairy root. Bars indicate SD (n = 3). (l) Effect of FK228 application on the performance of JACK and stable GmNFYA‐OE‐3 transgenic plants after 150 mM NaCl treatment for 3 days. Scale bar = 5 cm. (m) Relative electrolyte leakage and survival rates of JACK and stable GmNFYA‐transgenic plants with or without FK228 supply under salt‐stress condition. (n) Impact of FK228 on the histone acetylation level. The roots treated with different concentrations of FK228 for 16 h were harvested for Western blot assay. (o) Effect of FK228 application on the expression of salt‐responsive genes in roots of JACK. Asterisks indicate significant difference compared with the corresponding controls (*, P < 0,05; **, P < 0.01)
