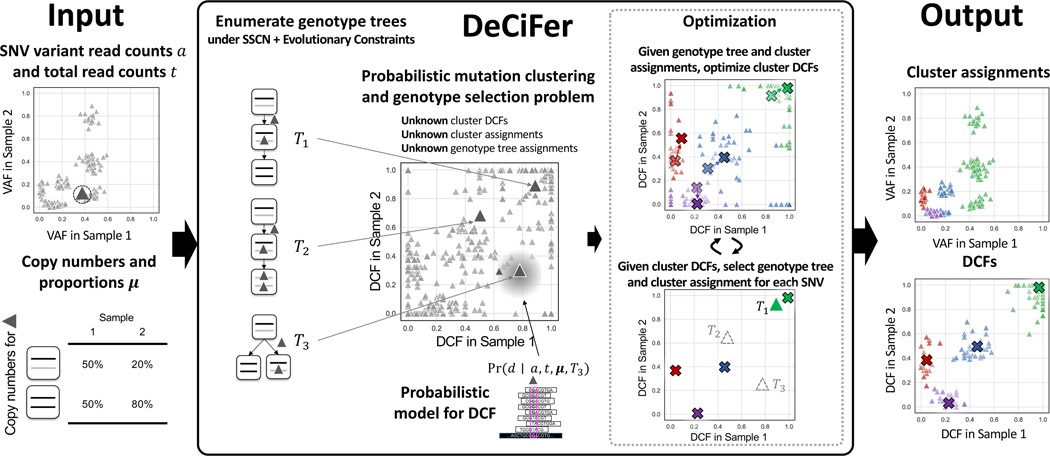
Figure 2: DeCiFer groups SNVs into clusters with shared evolutionary history, accounting for copy number aberrations and mutation loss.
(Left) Inputs to DeCiFer are variant a and total t read counts for SNVs identified in one or more tumor samples as well as the inferred copy numbers (x, y) and proportions μ(x, y) for each SNV locus. (Middle) DeCiFer examines possible genotype trees for each SNV conforming to the single split copy-number (SSCN) assumption and evolutionary constraints. Each genotype tree defines a transformation between the input data and a DCF value d in each sample. DeCiFer solves the probabilistic mutation clustering and genotype selection problem, simultaneously selecting a genotype tree for each SNV and clustering the DCF values determined by the selected genotype trees for all SNVs. DeCiFer uses a coordinate ascent approach, alternating between optimization of the DCF for each cluster given genotypes and cluster assignments followed by the selection of genotypes and cluster assignments given the DCF of each cluster. (right) Clusters output by DeCiFer often contain groups of SNVs with distinct VAFs (green points) but similar DCF values. These are a result of differing mutation multiplicities at SNV loci as well as loss of mutations in subpopulations of tumor cells.