FIGURE 5.
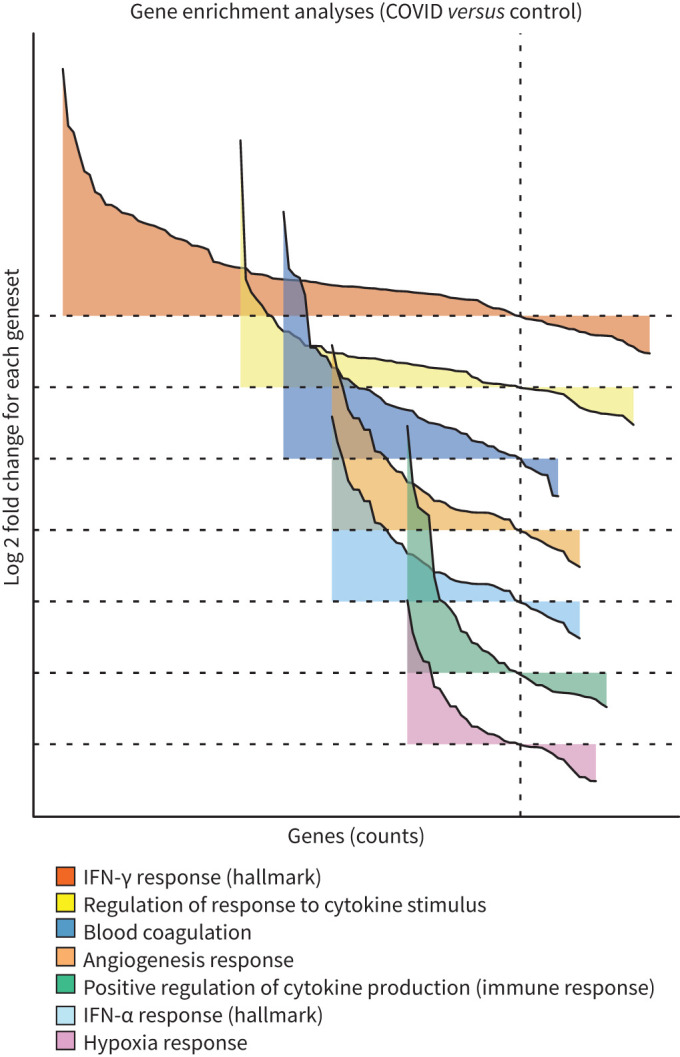
Gene set enrichment analysis (GSEA) reveals upregulated cytokine responses with accompanying coagulopathies in coronavirus disease 2019 (COVID-19) infection. GSEA for differentially expressed genes comparing COVID-19 and uninfected samples were conducted using the estimated log2 fold change and p-values with the distribution of significantly differentiated gene sets visualised as a function of the log2 fold change (logFC) and genes sorted by the logFC (waterfall plot). The gene sets visualised are custom gene sets for “angiogenesis response”, “blood coagulation”, “hypoxia” responses based on NanoString's nCounter PanCancer Progression Panel were identified by GSEA to be differentially upregulated in COVID-19 samples (table 2); Molecular Signatures Database (MSigDB) hallmark gene set for interferon (IFN)-α and IFN-γ and MSigDB Gene Ontology (GO) gene sets for “regulation of response to cytokine stimulus” and “positive regulation of cytokine production involved in immune response”. Gene sets ordered by gene counts. Refer to the comprehensive list in table 1. GSEA conducted using limma:fry with false discovery rate <0.05. The direction and relative change in expression are shown. Details of each gene set are provided in supplementary figures S4 and S5.
