Figure 6.
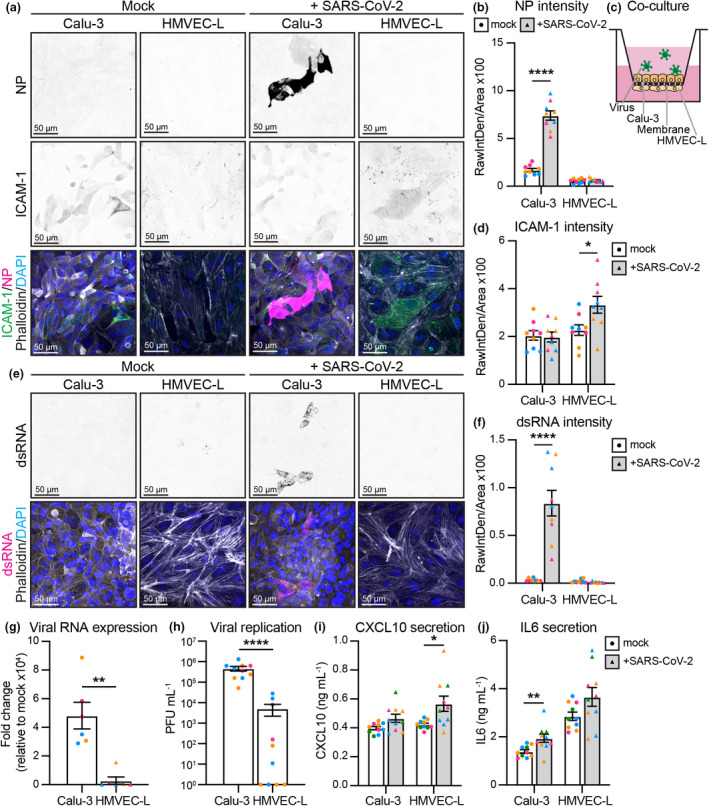
Co‐culture shows SARS‐CoV‐2 infection of Calu‐3 and inflammation of HMVEC‐L. (a) Representative immunofluorescence images of co‐cultured Calu‐3 and HMVEC‐L stained for nucleocapsid protein (NP) (shown as single channel in top panel) (magenta), ICAM‐1 (shown as single channel in middle panel) (green), Phalloidin (grey) and DAPI (blue) with mock or SARS‐CoV‐2 infection at 72 h after infection (2 × 106 PFU). Scale bar 50 µm. (b) Quantification of NP staining intensity in Calu‐3 and HMVEC‐L. n = 9 images from 3 independent experiments. (c) Schematic of Calu‐3 and HMVEC‐L co‐cultures on transwell membranes. (d) Quantification of ICAM‐1 staining intensity in Calu‐3 and HMVEC‐L. n = 9 images from 3 independent experiments. (e) Representative immunofluorescence images of co‐cultured Calu‐3 and HMVEC‐L stained for dsRNA (shown as single channel in top panel) (magenta), Phalloidin (grey) and DAPI (blue) with mock or SARS‐CoV‐2 infection at 72 h after infection. Scale bar 50 µm. (f) Quantification of dsRNA staining intensity in Calu‐3 and HMVEC‐L. n = 9 images from 3 independent experiments. (g) qPCR shows presence of viral RNA (MPRO) in SARS‐CoV‐2 infected Calu‐3 and HMVEC‐L co‐cultured cells, represented as fold change relative to mock infection at 72 h after infection. n = 3 independent experiments. (h) Viral replication shown as number of PFU mL−1 of supernatant from SARS‐CoV‐2 infected Calu‐3 and HMVEC‐L co‐cultured cells at 72 h after infection. n = 3 independent experiments. Measurement of cytokines with an AlphaLISA Immunoassay kit for (i) CXCL10 and (j) IL‐6 in the supernatant of Calu‐3 and HMVEC‐L co‐cultured cells with mock or SARS‐CoV‐2 infection at 72 h after infection. n = 4 independent experiments. Data are presented as mean ± s.e.m. with individual data points indicated and colour coded per independent experimental replicate. Statistical significance was determined using the Mann–Whitney U‐test between mock and + SARS‐CoV‐2 (b, d, f, g, h, i, j). *P < 0.05, **P < 0.01, ****P < 0.0001.
