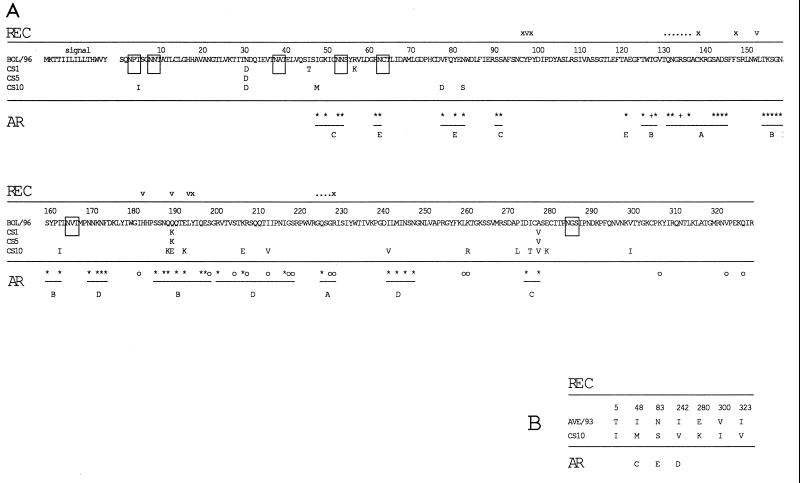
FIG. 2.
(A) Amino acid sequence alignment of HA1 regions of three field strains of the H3N8 subtype of equine influenza virus with the sequence of BOL/96, the most recently isolated Swedish strain. The sequence of CS9 differs from that of CS1 only at amino acid 42 of HA2 (alanine to leucine change) and has therefore not been included in the alignment. (B) Amino acid sequence alignment of HA1 of CS10 with HA1 of AVE/93, the most recently isolated Swedish representative of the European lineage of equine influenza virus subtype H3N8. Where present, amino acid differences in field strains are denoted below the BOL/96 and AVE/93 sequences. Possible N-linked glycosylation sites are boxed. The positions of amino acids involved in the receptor activity of HA are shown above the sequence alignment in the row named REC. v denotes an amino acid which makes up the receptor binding pocket structure, x denotes an amino acid involved in stabilizing the pocket, a dot denotes an amino acid which can interact with the receptor, ∗ denotes an amino acid found to vary in epidemic strains of human influenza virus subtype H3N2, and ° denotes an amino acid found to vary in laboratory-selected escape mutants. The positions of amino acids which have been identified as components of antigenic regions, and their corresponding antigenic regions, are shown below the sequence alignment on the row designated AR.