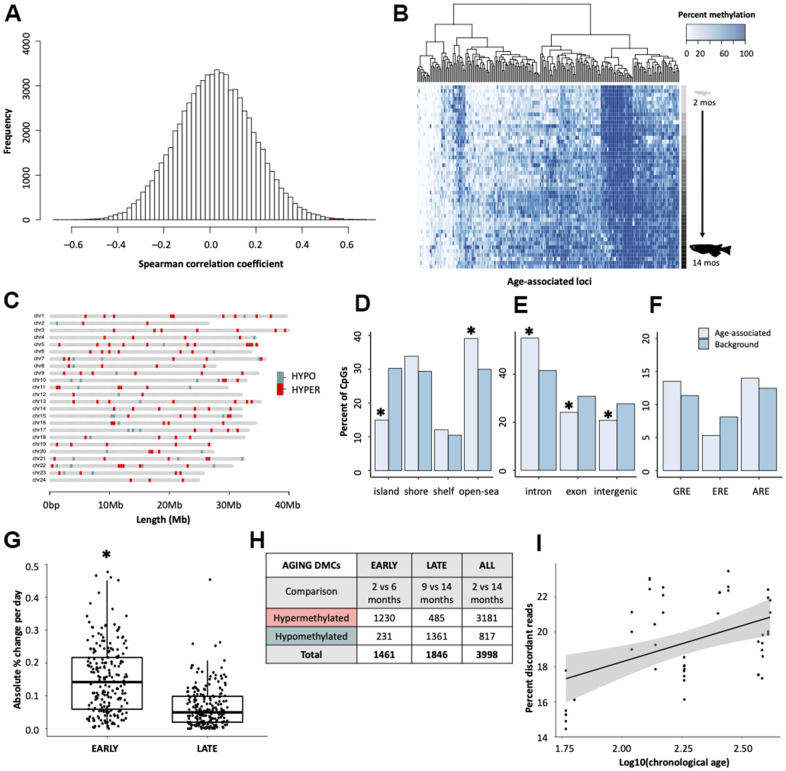
Figure 1.
Characterization of age-associated DNA methylation patterning in medaka hepatic tissue. (A) Histogram of correlation coefficients between methylation status and age in days. Hypermethylated cytosines are shown in red and hypomethylated in blue. (B) Heatmap of age-associated cytosines (n = 207). Age is specified by color intensity (2-month: light gray to 14-month: black). (C) Distribution of age-associated cytosines across the medaka genome. Cytosines that become hypermethylated with age are shown in red and those that become hypomethylated with age in blue. (D–F) Bar plots showing comparisons between age-associated cytosines (light blue) and background (dark blue) coverage of genomic features. (G) Comparison of the change in methylation during early- and late-life across age-associated cytosines. (H) Table showing differential methylation between early- and late-life. (I) Differences in the percent of reads which have discordant methylation across age groups.