ABSTRACT
The fungal zinc finger transcription factor NsdC is named after, and is best known for, its essential role in sexual reproduction (never in sexual development). In previous studies with Aspergillus nidulans, it was also shown to have roles in promotion of vegetative growth and suppression of asexual conidiation. In this study, the function of the nsdC homologue in the opportunistic human pathogen A. fumigatus was investigated. NsdC was again found to be essential for sexual development, with deletion of the nsdC gene in both MAT1-1 and MAT1-2 mating partners of a cross leading to complete loss of fertility. However, a functional copy of nsdC in one mating partner was sufficient to allow sexual reproduction. Deletion of nsdC also led to decreased vegetative growth and allowed conidiation in liquid cultures, again consistent with previous findings. However, NsdC in A. fumigatus was shown to have additional biological functions including response to calcium stress, correct organization of cell wall structure, and response to the cell wall stressors. Furthermore, virulence and host immune recognition were affected. Gene expression studies involving chromatin immunoprecipitation (ChIP) of RNA polymerase II (PolII) coupled to next-generation sequencing (Seq) revealed that deletion of nsdC resulted in changes in expression of over 620 genes under basal growth conditions. This demonstrated that this transcription factor mediates the activity of a wide variety of signaling and metabolic pathways and indicates that despite the naming of the gene, the promotion of sexual reproduction is just one among multiple roles of NsdC.
KEYWORDS: Aspergillus fumigatus, sexual cycle, transcription factor, cell wall integrity pathway, calcium signaling, transcription factors
INTRODUCTION
Aspergillus fumigatus is an opportunistic human fungal pathogen and the main causal agent of invasive aspergillosis, a life-threatening infection especially in immunocompromised patients (1). It is estimated that 300,000 infections occur per year worldwide with an associated mortality that could reach 95% (2, 3). The fungus is ubiquitous in the environment, and the infection is acquired mainly by inhalation of conidia present in the air (4). These conidia are produced on structures called conidiophores which are characteristic of asexual reproduction in Aspergillus species (5, 6). A. fumigatus can undergo both asexual and sexual reproductive cycles, but whereas asexual reproduction is observed in almost all aspergilli, sexual reproduction has been detected only in approximately 36% of Aspergillus species (7, 8).
The regulation of asexual reproduction involves several genes and pathways in A. fumigatus. A highly conserved pathway has been described as the central regulatory mechanism of asexual development in Aspergillus species such as A. fumigatus (6). It involves three key regulators that behave as a cascade activating each other in sequential order: BrlA, a transcription factor (TF) which is an essential activator; AbaA, responsible for hyphal differentiation into phialides; and WetA, important for conidial maturation (9–13). Accumulation of mRNA of the abaA and wetA genes was detected during asexual reproduction of A. fumigatus, while deletion of brlA impaired abaA and wetA transcript production (12). Further complementary regulatory mechanisms have also been reported to play a role in the genetic control of conidiation, such as the velvet family of negative regulator proteins (14), the FluG and FlbA-E upstream activators (11, 15), and members of the heterotrimeric G-protein signaling pathway (6, 11, 16, 17). Moreover, we have recently found that the mitogen-activated protein kinase MpkB is also important for conidiation and melanin production in A. fumigatus (18).
Despite evidence for cryptic sexual development in A. fumigatus having already been reported in the literature (19–21), it was not until 2009 that O’Gorman and colleagues were able to demonstrate the induction of a heterothallic (obligate outbreeding) sexual cycle under well-defined laboratory conditions (22). This involved formation of sexual fruiting bodies known as cleistothecia which at maturity contained sexual ascospores, with evidence provided for genetic recombination in the offspring. Sexual reproduction was found to require very specific environmental conditions not easily found in nature and incubation for up to 12 months. However, the subsequent identification of “supermater” strains (with enhanced sexual fertility) has facilitated the investigation of this process in a laboratory setting (23). The sexual cycle has provided an excellent tool not just for expanding basic genetic studies but also for investigating and explaining medically important processes such as the acquisition of azole resistance or differences in virulence in strains of A. fumigatus (24). The breeding system in A. fumigatus is governed by the presence of two mating-type (MAT) genes (MAT1-1 and MAT1-2) which encode alpha- and high-mobility group (HMG) domain transcription factors, respectively, and determine sexual identity of isolates (6, 20). Deletion of either of these genes resulted in a block in cleistothecium formation (25). Distribution of the compatible mating types has been reported to be near 1:1 in both regional and global populations of the fungus (20, 26).
Although a sexual cycle has only recently been described in A. fumigatus, Aspergillus nidulans has long been established as a model organism for studying processes controlling sexual development in ascomycete fungi. Beyond MAT genes, a wide list of over 70 sex-related genes has also been identified in A. nidulans, including genes with roles in light and nutrient sensing, signal transduction pathways, genes encoding transcription factors and other regulatory proteins, or even genes linked to endogenous physiological processes (27). Some of these findings have been confirmed to apply to A. fumigatus, such as the involvement of the transcription factor NsdD in hyphal fusion, necessary for heterokaryon formation (25). However, it is important to note that whereas A. fumigatus is a heterothallic species, in contrast A. nidulans is a homothallic (self-fertile) species and there is limited evidence that the regulation of sexual reproduction may differ slightly in homothallic versus heterothallic species (28).
The NsdC (never in sexual development) transcription factor was first identified in A. nidulans specifically due to its requirement for sexual reproduction. It encodes a fungus-specific C2H2-type zinc finger transcription factor, and its loss resulted in the lack of fruiting body formation, retarded vegetative growth, and hyperactive asexual sporulation (29). In the present work, we have characterized the homologous NsdC transcription factor from A. fumigatus, which was initially identified in a screen of transcription factor null mutants showing sensitivity when exposed to high concentrations of calcium (30). Besides calcium, we now report that the ΔnsdC strain exhibits increased sensitivity to cell wall-damaging agents. Furthermore, deletion of nsdC in both mating partners was found to completely abolish sexual development, while asexual conidiation was derepressed in liquid medium. Transmission electron microscopy, cell wall staining studies, and cell wall sugar content measurements showed that NsdC is involved in cell wall organization and composition, while genome-wide transcription profiling analysis (as measured by RNA polymerase II [PolII] occupancy) revealed NsdC has roles in several important biological responses. Finally, the ΔnsdC strain displayed a reduction in mortality and fungal burden in a neutropenic mouse model of invasive aspergillosis coupled with enhanced killing by macrophages. In summary, it is concluded that NsdC is important not only for sexual and asexual development but also for calcium tolerance and cell wall damage stress response by affecting cell wall composition and organization and has a role in virulence of A. fumigatus.
RESULTS
NsdC is important for calcium tolerance.
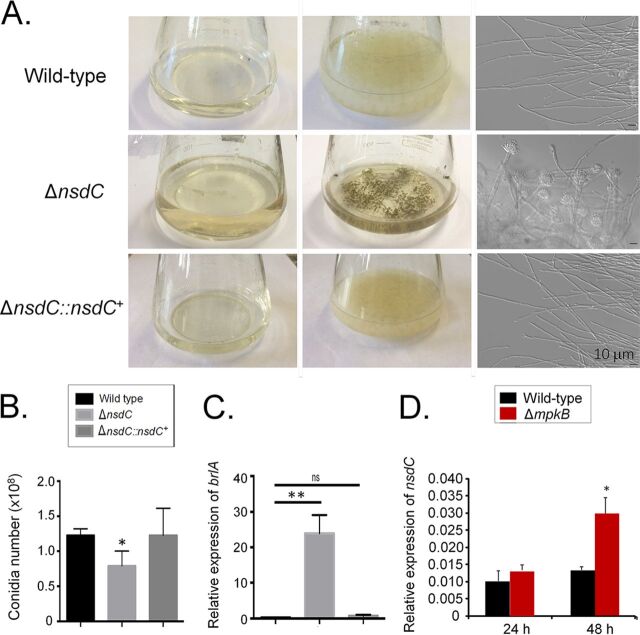
We previously identified, in a library of 395 A. fumigatus TF null mutants, nine null mutants that exhibited increased sensitivity to 500 mM calcium chloride (30). Among these TF mutants, we observed ΔnsdC (Afu7g03910), a putative homologue of A. nidulans NsdC (AN4263), a C2H2 zinc finger TF required for sexual development (29). In order to investigate the function of nsdC in A. fumigatus, the null mutant strain was here complemented by reinsertion of the wild-type gene and aspects of growth were compared between the deletion mutant and the complemented strain. The deletion of nsdC in A. fumigatus affected about 30 and 10% radial growth reduction on solid minimal medium (MM) and complete medium (yeast agar glucose [YAG]), respectively (Fig. 1A). There is about 20% growth reduction in liquid MM in 24-h growth compared to the wild type, but at 48 h growth is comparable to both wild-type and complemented strains (Fig. 1A, right panel). Interestingly, there are no growth differences in liquid YG medium for all three strains (Fig. 1A, right panel). The ΔnsdC mutant was more sensitive to calcium (CaCl2) (Fig. 1B) and exhibited enhanced resistance to the calcineurin activity inhibitor cyclosporine (Fig. 1C). We included these experiments with cyclosporine to see the relationship between calcineurin and NsdC. The ΔcalA mutant (null mutant for the calcineurin catalytic subunit) is also sensitive to calcium but resistant to cyclosporine (Fig. 1B and C). The ΔcrzA mutant is sensitive to calcium but as sensitive to cyclosporine as the wild-type strain (Fig. 1B and C). We have not observed differences between the morphology and growth of the wild-type and ΔnsdC germlings (Fig. 1D). NsdC-GFP (green fluorescent protein) under the control of the nsdC promoter (the cassette was homologously integrated in the nsdC locus) was constitutively present in the nucleus, and there was no clear effect on its translocation to the cytoplasm and/or degradation after exposure to high concentrations of CaCl2 or cyclosporine (Fig. 1E). Taken together, these results strongly suggest that NsdC is involved in the A. fumigatus calcium response pathway.
FIG 1.
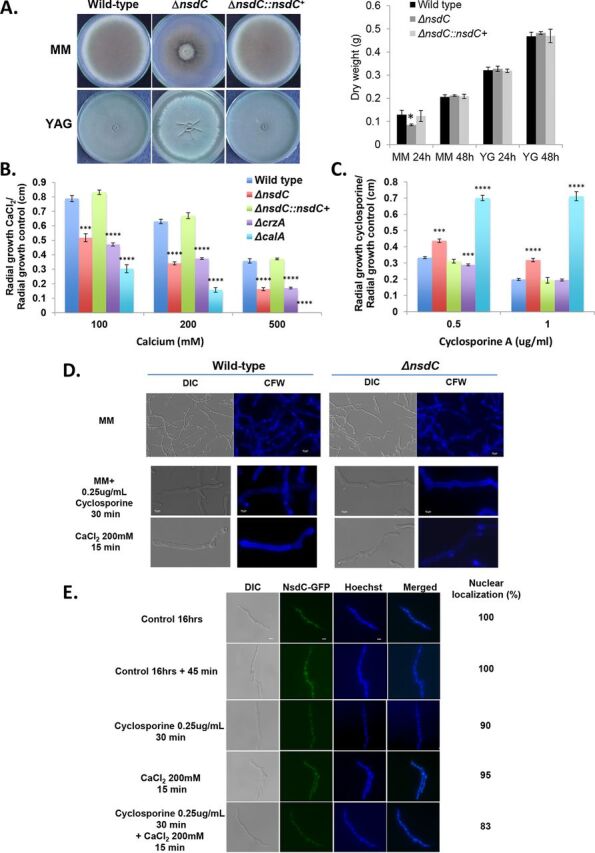
Growth phenotypes for ΔnsdC. (A to C) The wild-type, ΔnsdC, and ΔnsdC::nsdC+ strains were grown for 5 days on solid MM and YAG or for 2 days in liquid MM and YG at 37°C (A), MM + CaCl2 (B), and MM + cyclosporine (C). Results are expressed as radial growth of treatment/radial growth of control (centimeters) and are the average of three independent biological repetitions ± standard deviation. The statistical analysis was one-tailed, paired t test. P values: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Conidia of the wild-type and ΔnsdC strains were germinated for 16 h at 37°C in liquid MM and exposed or not to 0.25 μg/ml cyclosporine for 30 min or 200 mM CaCl2 for 15 min. Germlings were stained with calcofluor white (CFW). DIC, differential interference contrast. First row shows ×40 magnification while second and third rows show ×100 magnification. (E) NsdC-GFP germlings were grown for 16 h at 37°C in MM and exposed to 200 mM CaCl2 for 15 min or 0.25 μg/ml cyclosporine for 30 min followed by 200 mM CaCl2 for 15 min. About 100 germlings were counted in each treatment. Bar, 5 μm. Statistical analysis was performed using a one-way ANOVA comparing both ΔnsdC and ΔnsdC::nsdC+ strains to the wild type (****, P < 0.0001).
Generation of ΔnsdC mutants in high-fertility MAT1-1 and MAT1-2 mating-type backgrounds.
In the homothallic fungus Aspergillus nidulans, the nsdC gene is crucial for sexual development (29). Therefore, it was of interest to investigate whether the same is true for the heterothallic fungus A. fumigatus. Unfortunately, the MAT1-1 wild-type strain CEA17 (and its KU80 mutant and derivatives used elsewhere in the present study) was found to be sterile in preliminary crossing efforts, which made it difficult to assess the specific contribution of nsdC to sexual development in this background. To overcome this limitation, the nsdC gene was instead deleted in the high-fertility MAT1-1 wild-type strain 47-51 (AfIR974 [22]). As expected, some of the transformants displayed a condensed growth phenotype and green pigmentation of the mycelium on the regeneration plates, consistent with the phenotype observed with the ΔnsdC mutation in the CEA17 background (Fig. 1A), and nsdC deletion was confirmed in these transformants.
To test the fertility of ΔnsdC mutants with the 47-51 parental background, and to potentially generate ΔnsdC mutants with a MAT1-2 mating-type background, crosses were set up between the known fertile A. fumigatus MAT1-2 strains 47-55 and 47-107 and two representative ΔnsdC MAT1-1 mutants, 47-51ΔnsdC4 and 47-51ΔnsdC5 (see Table S1 at https://doi.org/10.6084/m9.figshare.12931754.v3). All crosses were found to produce cleistothecia and viable ascospores (see Table S2 at https://doi.org/10.6084/m9.figshare.12931754.v3). This clearly demonstrated that nsdC is not essential for fertility in crosses with a heterothallic wild-type mating partner, at least when nsdC is deleted in the MAT1-1 genetic background. The ΔnsdC mutants appeared to show slightly lower fertility than the 47-51 parent when crossed to isolate 47-55, but this reduction was not statistically significant (F = 1.369; P value = 0.2820). When ascospores were plated on a medium without pyrithiamine, both wild-type and ΔnsdC mutant phenotypes were observed, whereas only the ΔnsdC phenotype occurred on plates with pyrithiamine (Fig. 2A to D). This was consistent with the use of the pyrithiamine resistance cassette in nsdC deletion. Ascospore offspring from crosses between 47-51ΔnsdC5 and 47-107 and between 47-51ΔnsdC5 and 47-55 were then randomly selected from the pyrithiamine plate and analyzed for their mating type (Fig. 2A to D). These ΔnsdC progeny showed an approximate 1:1 ratio of MAT1-1 to MAT1-2 genotypes, confirming that sexual recombination had occurred as demonstrated by the appearance of the novel ΔnsdC MAT1-2 genotypes (Fig. 2A to D; see also Table S1 at https://doi.org/10.6084/m9.figshare.12931754.v3).
FIG 2.
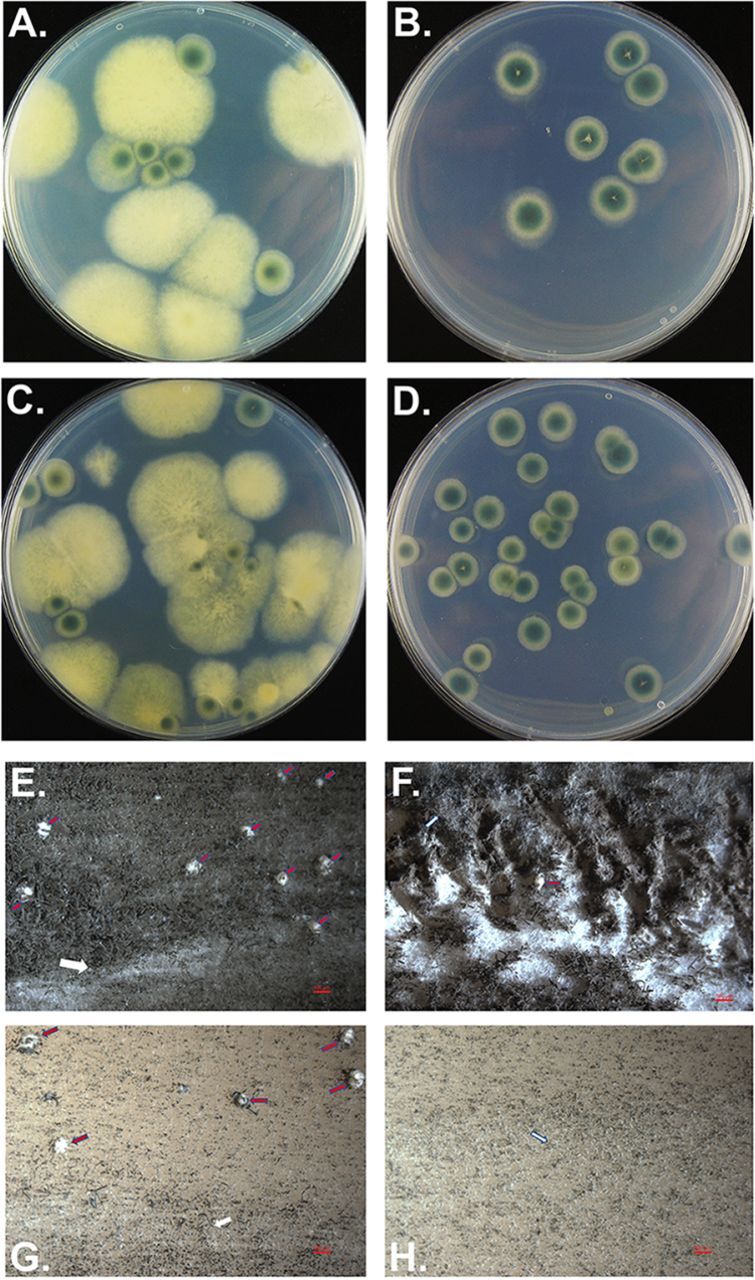
Sexual crosses of the ΔnsdC mutant. Appearance of sexual progeny from crosses using 47-51ΔnsdC (5) as a mating partner, in which nsdC was deleted by replacement with a pyrithiamine resistance cassette. (A and B) Offspring of the cross 47-51ΔnsdC (5) × 47-55. (C and D) Offspring of the cross 47-51 ΔnsdC (5) × 47-107. Ascospore progeny were spread plated onto either Glucose 50, Gln 10 (A and C) or Glucose 50, Gln 10, PT (pyrithiamine) 0.1 (B and D) medium; see Materials and Methods for details. All pyrithiamine-resistant (at 0.1 μg/ml PT) offspring (B and C) show the ΔnsdC phenotype. (E to H) Cleistothecia formed by representative mating partners of A. fumigatus on oatmeal agar following incubation for 4 months in darkness at 30°C. Images photographed after “hoovering” of plates as appropriate to allow clearer visualization of cleistothecia. Red arrows indicate cleistothecia, and white arrows indicate the barrage zone where two mating types of A. fumigatus strains met. Bar, 500 μm. (E) Cleistothecia formed in a cross between strains 47-267 (MAT1-1) and 47-55 (MAT1-2). (F) Cleistothecia formed in a cross between 47-267 and ΔnsdC-a (MAT1-2). (G) Cleistothecia formed by 47-55 and ΔnsdC-b (MAT1-1). (H) Absence of cleistothecia in a cross between ΔnsdC-a and ΔnsdC-b.
PCR analysis of these transformants further confirmed the successful modification of the nsdC gene in these transformants (see Fig. S1 at https://doi.org/10.6084/m9.figshare.12931754.v3).
Impact of both MAT1-1 and MAT1-2 nsdC deletion on sexual crossing of A. fumigatus.
Since a MAT1-1 ΔnsdC strain was fertile when crossed with a highly fertile MAT1-2 strain, we were interested to see whether this also held true for a ΔnsdC mutant with a MAT1-2 mating-type background. Furthermore, we investigated the result of crossing of eight additional ΔnsdC strains, comprised of two MAT1-1 and two MAT1-2 strains from progeny of each of crosses between 47-51-ΔnsdC5 and 47-55 or 47-51-ΔnsdC5 and 47-107 (see Table S2 at https://doi.org/10.6084/m9.figshare.12931754.v3).
As shown in Table 1 and Fig. 2E to H, cleistothecia were formed in all control crosses between MAT1-1 and MAT1-2 wild-type isolates, and also in almost all crosses when at least one mating partner contained a functional copy of the nsdC gene, irrespective of whether it was a MAT1-1 or MAT1-2 partner that contained the functional copy. The only exceptions were crosses 47-267 × ΔnsdC-w and 47-107 × ΔnsdC-y, which failed to form cleistothecia, but importantly strains ΔnsdC-w and ΔnsdC-y formed cleistothecia with the alternative mating partners 47-51 and 47-55, respectively. In contrast, deletion of nsdC in both mating partners resulted in a complete lack of the formation of cleistothecia in all 16 test crosses, despite incubation for an extended period of 6 months and the formation of a barrage zone between the partners (Table 1; Fig. 2E to H). Therefore, it can be concluded that whereas gene activation by nsdC in one mating partner is sufficient to initiate and allow completion of the sexual cycle, the absence of nsdC in both mating partners instead totally abolishes sexual development.
TABLE 1.
Formation of cleistothecia in crosses between various MAT1-1 A. fumigatus wild-type and ΔnsdC strains and various MAT1-2 A. fumigatus wild-type and ΔnsdC strainsa
| MAT1-2 strain |
MAT1-1 strain |
|||||
|---|---|---|---|---|---|---|
| 47-51 | 47-267 | ΔnsdC-b | ΔnsdC-z | ΔnsdC-y | ΔnsdC-d | |
| 47-55 | 7.0 ± 5.7 | 29.7 ± 8.0 | 13.8 ± 9.2 | 2.8 ± 2.1 | 7.8 ± 7.6 | 8.3 ± 11.0 |
| 47-107 | 14.2 ± 8.7 | 0.3 ± 0.5 | 4.0 ± 4.7 | 0.5 ± 0.6 | 0 | 0.5 ± 1.0 |
| ΔnsdC-a | 0.5 ± 1.0 | 1.5 ± 1.7 | 0 | 0 | 0 | 0 |
| ΔnsdC-c | 5.5 ± 5.9 | 2.7 ± 5.5 | 0 | 0 | 0 | 0 |
| ΔnsdC-x | 35.0 ± 21.8 | 21.2 ± 8.1 | 0 | 0 | 0 | 0 |
| ΔnsdC-w | 40.7 ± 18.9 | 0 | 0 | 0 | 0 | 0 |
Numbers indicate average numbers of cleistothecia per 9-cm crossing plate ± SEM.
NsdC is involved in the control of asexual sporulation.
The ΔnsdC strain displayed deregulation in initiation of conidiation in liquid MM, with conidiophores and conidia being produced, unlike the control wild-type parent and complemented strains (Fig. 3A). However, a significant reduction in the number of conidia produced on solid MM by the ΔnsdC strain was observed in comparison to the control strains (Fig. 3B). Previous reports described that the transcription factor BrlA is important for the activation of conidial development in A. fumigatus (11). RT-qPCR experiments showed that brlA expression in the ΔnsdC strain was 25 times higher in liquid MM than in the wild-type and complemented strains (Fig. 3C). We have previously observed that the mitogen-activated protein (MAP) kinase MpkB plays an important role in the negative control of conidiation in liquid medium (18). The accumulation of nsdC mRNA was dependent on MpkB since there was a 2-fold increase in the nsdC mRNA transcripts in the ΔmpkB strain compared with the wild-type strain after 48-h growth in liquid medium (Fig. 3D). These results indicate that NsdC is involved in suppression of conidiation in liquid culture but also has a role in promoting hyphal vegetative growth and concomitant asexual sporulation on solid media (as per Fig. 1A) and that nsdC expression is, at least to some degree, dependent on MpkB.
FIG 3.
The A. fumigatus nsdC null mutant produces conidiophores in liquid cultures with agitation. (A) Cultures (center column) and separated supernatants from the same cultures (left column) were grown with agitation for 5 days at 37°C. DIC microscopic analysis revealed the presence of conidia and conidiophores in the ΔnsdC cultures (right column). (B) Number of conidia in the wild-type, ΔnsdC, and ΔnsdC::nsdC+ strains grown for 5 days at 37°C on MM. (C) The wild-type, ΔnsdC, and ΔnsdC::nsdC+ strains were grown for 24 h at 37°C in MM. Gene expression was obtained for brlA and was normalized by using tubA (Afu1g10910). (D) The wild-type and ΔmpkB strains were grown for 24 or 48 h at 37°C in MM. Gene expression was obtained for nsdC and was normalized by using tubA (Afu1g10910). Error bars represent standard deviations of the average of three independent biological repetitions (each with 2 technical repetitions). Statistical analysis was performed using a one-way ANOVA compared to the wild-type condition (*, P < 0.05; **, P < 0.01; ns, not significant).
NsdC participates in organization of the cell wall.
Although 1.2 M sorbitol improved ΔnsdC mutant radial growth (Fig. 4A), the mutant strain was more sensitive to the chitin synthase inhibitor nikkomycin Z (Fig. 4B) and other cell wall-damaging agents, including Congo red (CR), calcofluor white (CFW), and caspofungin than were the wild-type and complemented strains (Fig. 4C to E). Increased sensitivity to cell wall stressors suggested the ΔnsdC mutant had an altered cell wall composition. Cell wall stains and fluorescently labeled lectins were therefore used to identify differences in the ability to detect (i.e., exposure of) several carbohydrates on the cell wall surface, for the wild-type, ΔnsdC, and complemented strains. These included (i) WGA (wheat germ agglutinin)-FITC (fluorescein isothiocyanate), recognizing surface-exposed glucosamine (Glc); (ii) SBA (soybean agglutinin)-FITC, which binds preferentially to oligosaccharide structures with terminal α- or β-linked N-acetylgalactosamine (GalNAc) and to galactose residues which are important for recognizing galactosaminogalactan (GAG); (iii) ConA (concanavalin A)-FITC, which recognizes α-linked mannose; (iv) CFW (recognizing chitin); and (iv) soluble dectin-1 staining (recognizing β-glucans). Despite finding no differences in the exposed Glc among all strains (Fig. 4F), the ΔnsdC mutant had about a 2- to 3-fold reduction in the exposure of GalNAc, α-linked mannose, chitin, and β-1,3-glucan compared to the wild-type and complemented strains (Fig. 4G to J). All the three strains have about the same amount of total carbohydrates in their cell wall (Fig. 4K); however, the mutant strain displayed lower exposure of the cell wall sugar components glucosamine, glucose, and NAG (N-acetylglucosamine) but increased mannose and galactose relative to the wild-type and complemented strains (Fig. 4L). Interestingly, transmission electron microscopy (TEM) experiments showed that the cell wall of the ΔnsdC mutant was about 5-fold thicker than those of the wild-type and complemented strains (Fig. 4M and N; see also Table S3 at https://doi.org/10.6084/m9.figshare.12931754.v3). Therefore, NsdC influences fungal cell wall composition organization and structure.
FIG 4.
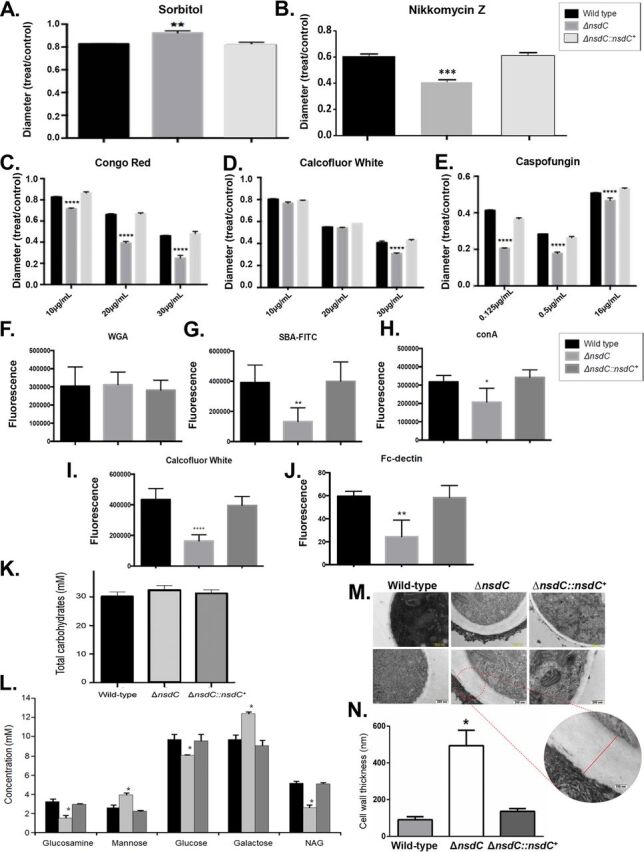
A. fumigatus NsdC affects the cell wall organization and structure. (A to E) The wild-type, ΔnsdC, and ΔnsdC::nsdC+ strains were grown for 5 days at 37°C in MM + sorbitol at 1.2 M (A), MM + nikkomycin Z at 100 μM (B), MM + Congo red up to 30 μg/ml (C), MM + calcofluor white up to 30 μg/ml (D), and MM + caspofungin up to 16 μg/ml (E). Error bars represent standard deviations of the average of three independent biological repetitions. Statistical analysis was performed using a one-way ANOVA compared to the wild-type condition (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (F to J) Detection of sugar exposed on the cell wall. Conidia were cultured in liquid MM until hyphae were produced, UV killed, and stained with WGA (wheat germ agglutinin)-FITC (recognizing surface-exposed glucosamine [Glc]) (F), SBA (soybean agglutinin)-FITC (preferentially binds to oligosaccharide structures with terminal α- or β-linked N-acetylgalactosamine [GalNAc] and, to a lesser extent, galactose residues—important for recognizing galactosaminogalactan [GAG]) (G), ConA (concanavalin A)-FITC (recognizes α-linked mannose) (H), CFW (recognizing chitin) (I), and soluble dectin-1 staining (recognizing β-glucans) (J). (K) Total carbohydrate concentration in the cell walls of the wild-type, ΔnsdC, and ΔnsdC::nsdC+ strains. (L) Sugar concentrations in the cell walls as determined by high-performance liquid chromatography (HPLC), in mycelial extracts of the A. fumigatus strains when grown for 16 h in MM at 37°C. Experiments were performed in triplicate, and results are displayed as mean values with standard deviations (one-way ANOVA followed by Tukey’s post hoc test [*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001]). (M) Transmission electron microscopy of mycelial sections of A. fumigatus wild-type, ΔnsdC, and ΔnsdC::nsdC+ strains following growth for 24 h in MM at 37°C. Inset shows a magnification of the ΔnsdC mutant cell wall. (N) The cell wall thickness (nanometers) of 100 sections of different hyphal germlings (average from 4 sections per germling) was measured. Standard deviations present the average from the 100 measurements, and statistical differences were evaluated by using one-way analysis of variance (ANOVA) and Tukey’s post hoc test (*, P < 0.05).
Chromatin immunoprecipitation of RNA polymerase II coupled to next-generation sequencing (PolII ChIP-Seq) for ΔnsdC.
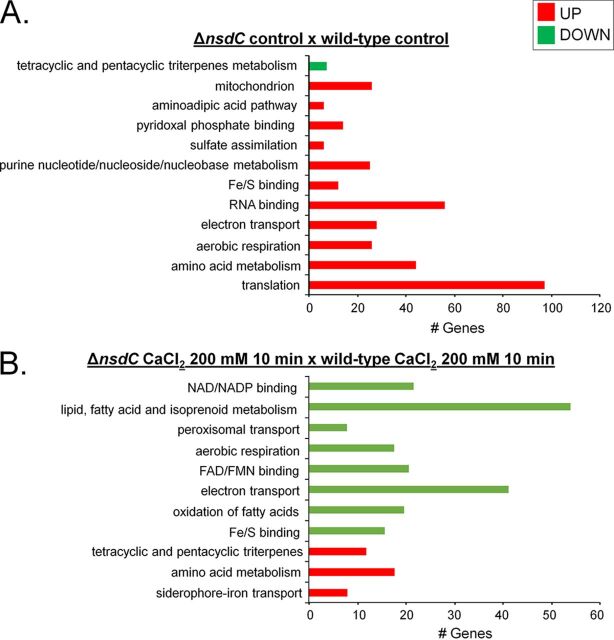
Considering that NsdC has many different functions besides its involvement in the sexual process, we decided to evaluate the transcriptional impact of the lack of nsdC after growth in MM for 16 h in the absence and presence of CaCl2. To investigate this, we used PolII ChIP-Seq with a corresponding wild-type strain and the ΔnsdC mutant. Considering that RNA PolII recruitment is related to transcriptional activity, a corresponding increase and decrease in RNA PolII occupancy indicate upregulation and downregulation of gene expression, respectively (31, 32). In the ΔnsdC strain compared with the wild type, we identified an increase in RNA PolII occupancy of 805 genes and reduction of PolII occupancy of 120 genes (log2FC [fold change] ≥ 1.0 and ≤ −1.0; false-discovery rate [FDR] of 0.05; see Table S4 at https://doi.org/10.6084/m9.figshare.12931754.v3). FunCat (https://elbe.hki-jena.de/fungifun/fungifun.php) enrichment analyses of the upregulated genes demonstrated an increased PolII occupancy for those encoding proteins involved in translation, mitochondrion, aerobic respiration, electron transport, and amino acid metabolism (Fig. 5A). FunCat categorization of the downregulated genes allowed us to identify only a single category of genes, in this case encoding proteins involved in tetracyclic and pentacyclic triterpene (cholesterin, steroid, and hopanoid) metabolism (Fig. 5A).
FIG 5.
A summary of the categorization terms of overrepresented up- or downregulated (adjusted P value < 0.05) genes after PolII ChIP-Seq for the ΔnsdC strain relative to the parental control. FunCat (https://elbe.hki-jena.de/fungifun/fungifun.php) enrichment analyses for the ΔnsdC control × wild-type control (A) and ΔnsdC CaCl2 (200 mM, 10 min) × wild-type CaCl2 (200 mM, 10 min) (B). FAD/FMN, flavin adenine dinucleotide/flavin mononucleotide.
The same method of transcriptional profiling was then used to compare gene expressions of the ΔnsdC and wild-type strains when both were exposed to 200 mM CaCl2 for 10 min. An increased RNA PolII occupancy was identified for 310 genes together with a reduction of PolII occupancy of 623 genes (log2FC ≥ 1.0 and ≤ −1.0; FDR of 0.05; see Table S4 at https://doi.org/10.6084/m9.figshare.12931754.v3). FunCat (https://elbe.hki-jena.de/fungifun/fungifun.php) enrichment analyses demonstrated an increased PolII occupancy of genes encoding siderophore-iron transport and amino acid and tetracyclic and pentacyclic triterpene (cholesterin, steroid, and hopanoid) metabolism (Fig. 5B). However, we have not observed any iron deficiency in the ΔnsdC strain radial growth (see Fig. S2 at https://doi.org/10.6084/m9.figshare.12931754.v3). FunCat categorization of the downregulated genes allowed us to identify proteins involved in Fe/S binding; electron transport; aerobic respiration; lipid, fatty acid, and isoprenoid metabolism; peroxisomal transport; amino acid metabolism; mitochondrial transport; and siderophore-iron transport (Fig. 5B).
We identified at least five genes encoding proteins involved in the sexual process as modulated under both transcriptional conditions (absence or presence of CaCl2 [Fig. 5A]; also see Table S4 at https://doi.org/10.6084/m9.figshare.12931754.v3): (i) rosA (AFUA_4g09710), overexpressed in the presence of CaCl2, a repressor of sexual development in A. nidulans (33); (ii) csnC (AFUA_2G07340), overexpressed in the absence of CaCl2, an A. nidulans ortholog that has a role in cleistothecium development and COP9 signalosome localization (34); (iii) osaA (AFUA_3g09640), overexpressed in the presence of CaCl2, encoding a protein with a WOPR domain involved in regulation of sexual development in A. nidulans (35); (iv) nsdD (AFUA_3g13870), overexpressed both in the absence and in the presence of CaCl2, encoding a GATA-type transcriptional activator and required during an early stage of mating (36); and (v) imeB (AFUA_2g13140), overexpressed in the presence of CaCl2, a serine/threonine protein kinase involved in the inhibition of sexual development in A. nidulans (37).
There are several genes encoding proteins involved in the asexual conidiation process that are overexpressed in the absence of CaCl2 (Fig. 6A; also see Table S4 at https://doi.org/10.6084/m9.figshare.12931754.v3), such as flbD (AFUA_1G03210, a Myb family transcription factor whose A. nidulans ortholog plays a role in conidiophore development [38]) and SfgA (AFUA_5G02800, a C6 transcription factor that has a role in negative regulation of conidium formation [39]), and in the presence of CaCl2, such as flbB (AFUA_2g16060, a Bzip developmental regulator involved in A. nidulans asexual development [40]), flbC (AFUA_2g13770, a C2H2 finger domain protein involved in asexual development in A. nidulans [40]), and vapA (AFUA_5g11190, a component of the plasma membrane-associated VapA-VipC-VapB methyltransferase complex that controls A. nidulans differentiation [41]).
FIG 6.
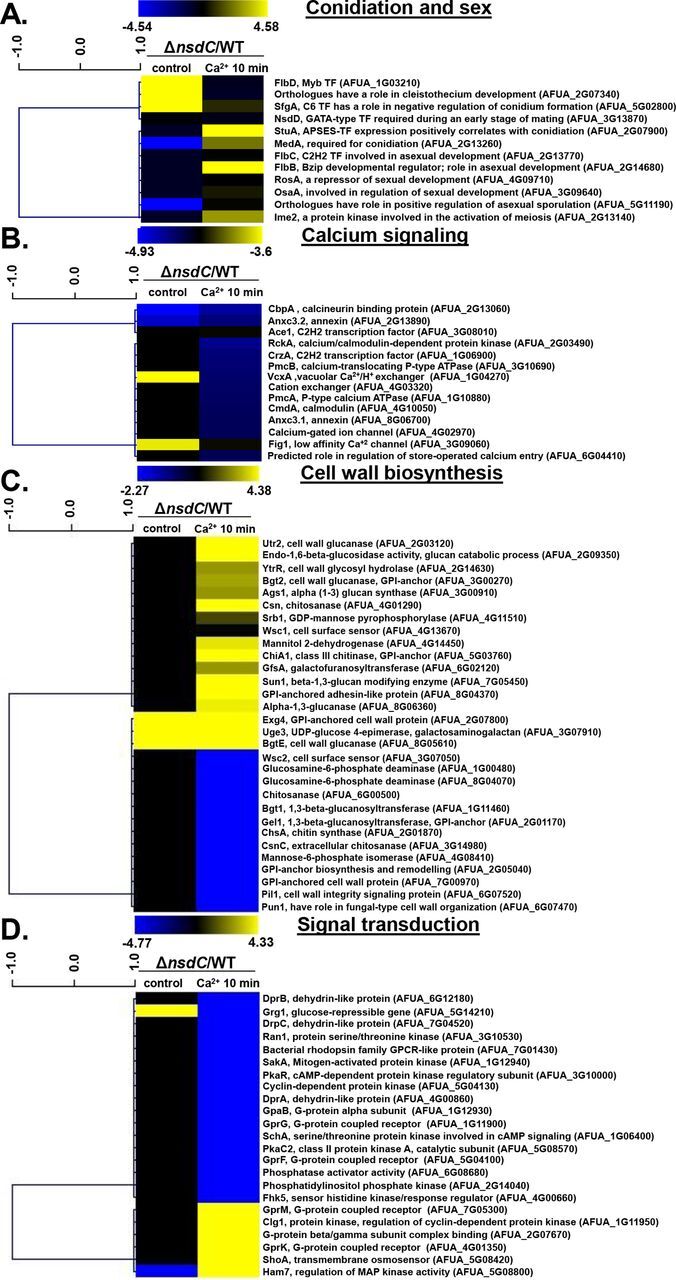
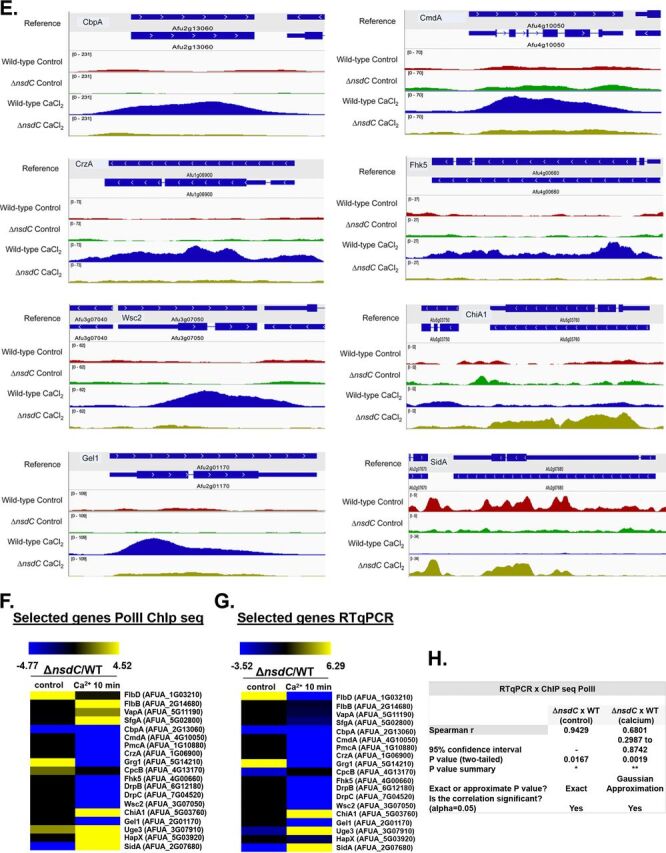
Independent validation of selected genes identified as modulated in PolII ChIP by RT-qPCR. (A to D) The results show the PolII occupancy for selected annotated genes encoding proteins putatively involved in conidiation and sex (A), calcium signaling (B), cell wall biosynthesis (C), and signal transduction (D). The ΔnsdC strain is shown as the deletion strain versus the equivalent wild-type (WT) strain for control and 10-min time point (the mutant values have been normalized to the basal level of expression of each gene before stress, i.e., expression ratios are being compared: the ΔnsdC control divided by the wild-type control and the ΔnsdC mutant 10 min versus time zero divided by the wild-type 10 min versus time zero). Hierarchical clustering was performed in MeV (http://mev.tm4.org/), using Spearman correlation with complete linkage clustering. (E) PolII ChIP-Seq Integrative Genomics Viewer (IGV; http://software.broadinstitute.org/software/igv/download) screenshot for selected genes from panels F and G. (F) Heat map for the PolII occupancy of 19 selected genes involved in conidiation and sex, calcium signaling, cell wall biosynthesis, and signal transduction. (G) Heat map of RT-qPCR for the same 19 selected genes from panel F. The wild-type and ΔnsdC mutant strains were grown for 20 h at 37°C and transferred to 200 mM CaCl2 for 0 and 10 min. Gene expression was normalized using cofA (Afu5g10570). Standard deviations present the average of three independent biological repetitions (each with 2 technical repetitions). (H) The expression of these 19 genes as measured by RT-qPCR showed a high level of correlation with the PolII ChIP-Seq data (Spearman correlation from 0.6801 to 0.9429).
Taken together, these results indicate that NsdC is important for amino acid, iron, and lipid metabolism as well as conidiation, impacting several biological responses under basal conditions and upon exposure to calcium stress.
Independent validation of NsdC influence on transcriptional regulation of calcium response.
RT-qPCR experiments validated the PolII ChIP-Seq results for the majority of the 19 selected genes from conidiation and sex, calcium signaling, cell wall, and signal transduction responses (Fig. 6A to G; see also Table S4 and Fig. S5 at https://doi.org/10.6084/m9.figshare.12931754.v3). The cofA (Afu5g10570) gene, which encodes the actin-binding protein cofilin, was used as a normalizer due to its consistent expression in all strains during calcium stress (30). The expression of these 19 genes showed a high level of correlation with the PolII ChIP-Seq data (Spearman correlation from 0.6801 to 0.9429 [Fig. 6H]). These results validate that NsdC has functions in modulating the response to calcium stress, affecting directly or indirectly the expression of genes involved in conidiation, sex, signal transduction, and cell wall biosynthesis and/or remodeling.
The A. fumigatus ΔnsdC mutant has attenuated virulence in immunodeficient mice.
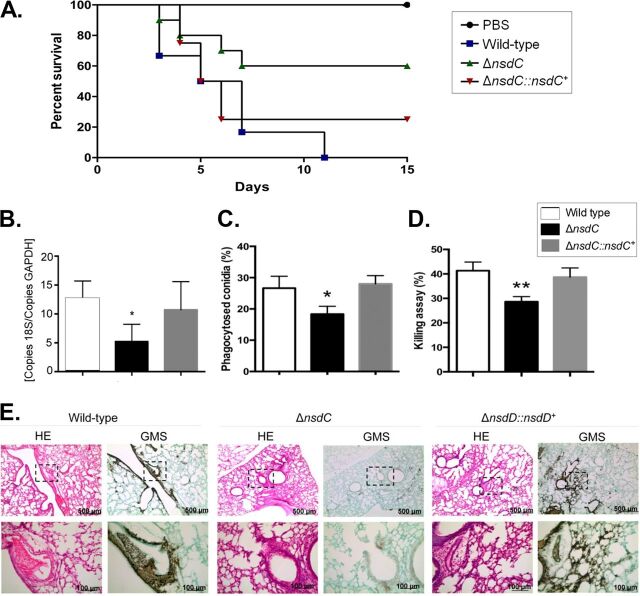
In the leukopenic BALB/c murine model of invasive pulmonary aspergillosis, wild-type and ΔnsdC::nsdC+ exposure resulted in 100 and 80% mortality at 11 and 15 days postinoculation, respectively (Fig. 7A). In contrast, the ΔnsdC mutant caused a reduced mortality of only 40% at 15 days postinoculation, which was statistically different from the wild-type and ΔnsdC::nsdC+ strains (Mantel-Cox and Gehan-Brestow-Wilcoxon tests; P values < 0.05 [Fig. 7A]). Additionally, fungal burden in lungs of mice was measured by qPCR and revealed a lower presence of ΔnsdC DNA than of that of the wild-type and complemented ΔnsdC::nsdC+ strains (Fig. 7B). These data strongly indicated that the lack of NsdC in A. fumigatus caused a significant reduction in virulence in immunodeficient mice and also indicated that the ΔnsdC strain might be more sensitive to macrophage killing.
FIG 7.
The A. fumigatus ΔnsdC strain has attenuated virulence in neutropenic mice. (A) Comparative analysis of wild-type, ΔnsdC, and ΔnsdC::nsdC+ strains in a neutropenic murine model of pulmonary aspergillosis. Mice in groups of 10 per strain were inoculated intranasally with a 20-μl suspension of conidia at a dose of 105. PBS, phosphate-buffered saline. The statistical significance of comparative survival values was calculated with the Prism statistical analysis package by using log rank (Mantel-Cox) and Gehan-Brestow-Wilcoxon tests. (B) Fungal burden was determined 48 h postinfection by real-time qPCR based on the 18S rRNA gene of A. fumigatus and an intronic region of the mouse GAPDH gene. Fungal and mouse DNA quantities were obtained from the CT values from an appropriate standard curve. Fungal burden was determined through the ratio between nanograms of fungal DNA and milligrams of mouse DNA. The results are the means (± standard deviations) from five lungs for each treatment. Statistical analysis was performed by using the t test (*, P < 0.05). (C and D) Fungal killing capacity of mouse bone marrow-derived macrophages (BMDMs) assessed as CFU of surviving A. fumigatus strains (wild type, ΔnsdC, or ΔnscD::nscD+ mutant) after 4 h of infection. The percent killing was calculated from CFU remaining compared with control samples without macrophages. The results are the means (± standard deviation) of three independent biological repetitions. Statistical analysis was performed using a one-way ANOVA compared to the wild-type condition (*, P < 0.05; **, P < 0.01). (E) Histopathology of mice infected with A. fumigatus wild-type or ΔnsdC or ΔnscD::nscD+ mutant strain. GMS (Grocott’s methenamine silver) and HE (hematoxylin and eosin) staining of lung sections representative of infections. The boxed area from the top row is magnified in the bottom row. Bars, 100 and 500 μm.
Histopathological examination revealed that at 72 h postinfection mouse lungs infiltrated with the ΔnsdC strain showed no sign of fungal burden. In contrast, mice infected with the wild-type or the ΔnsdC::nsdC+ strain contained multiple foci of invasive hyphal growth, which penetrated the pulmonary epithelium in major airways (Fig. 7E). These data strongly indicated that the lack of NsdC in A. fumigatus caused a significant reduction in virulence in immunodeficient mice and also indicated that the ΔnsdC strain might be more sensitive to macrophage killing.
Since loss of the nsdC gene produced alterations in cell wall composition (Fig. 4), we hypothesized that it could influence the immune host response and virulence. Macrophages contribute to innate immunity, fungal clearance, and the generation of a proinflammatory response during A. fumigatus exposure to the microbe (42). Thus, the capacity of bone marrow-derived macrophages (BMDMs) to phagocytose and kill the wild-type, ΔnsdC, and ΔnsdC::nsdC+ conidia was assessed. The nsdC mutant conidia presented lower killing rates by BMDMs than did the wild-type and nsdC::nsdC+ conidia (Fig. 7C and D). These results indicate that the nsdC mutant was less susceptible to macrophage killing.
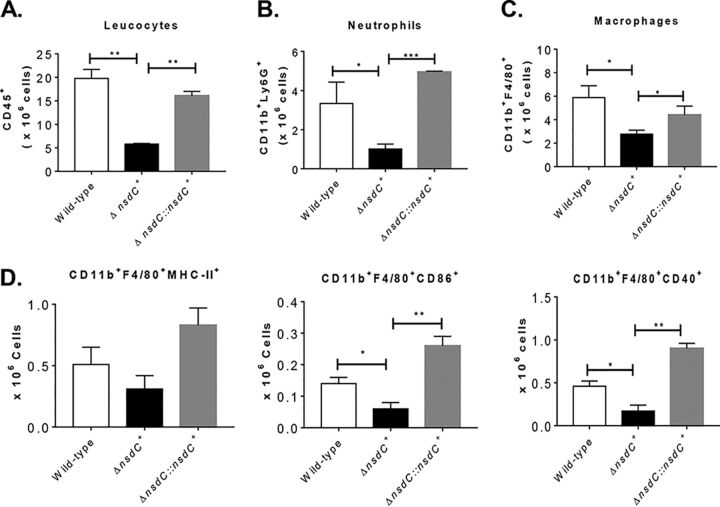
In addition, experiments with immunocompetent C57BL/6 mice were performed to assess the ability of the ΔnsdC strain to generate an inflammatory response. The immunocompetent mice were inoculated with 5 × 107 conidia of wild-type, ΔnsdC, and ΔnsdC::nsdC+ strains by the intratracheal (i.t.) route. After 72 h of inoculation, the lung-infiltrating leukocytes were obtained and analyzed for the expression of surface molecules of leukocytes by flow cytometry. As shown in Fig. 8A to D and also in Fig. S3 at https://doi.org/10.6084/m9.figshare.12931754.v3, lower numbers of total leukocytes (CD45+ cells), neutrophils, and macrophages were found in the lungs of mice exposed to ΔnsdC mutant strains compared with wild-type and ΔnsdC::nsdC+ conidium inoculation. Similarly, fewer numbers of inflammatory macrophage cells (CD11b+ F4/80+ major histocompatibility complex class II [MHC-II+], CD11b+ F4/80+ CD86+, and CD11b+ F4/80+ CD40+, respectively) were detected in the lungs of mice inoculated with ΔnsdC mutant strains compared with wild-type and ΔnsdC::nsdC controls. These data strongly indicate that the lack of NsdC in A. fumigatus caused a significant reduction in virulence in immunodeficient and immunocompetent mice and also indicated that the ΔnsdC strain was more sensitive to macrophage killing.
FIG 8.
ΔnsdC determines impaired leukocyte influx to the lungs of immunocompetent C57BL/6 mice. Mice were inoculated with 5 × 107 conidia of A. fumigatus strains by i.t. route, and cell phenotypes were determined after 72 h of infection. Lungs from mouse groups (n = 3) were excised and digested enzymatically. Cell suspensions were obtained and stained as described in Materials and Methods. The stained cells were analyzed immediately on a FACSLyrics flow cytometer with gating of granulocytes, as judged from forward scatter (FSC) and side scatter (SSC). The number of total CD45+ leukocytes (A), CD11b+ F4/80− Ly6G+ neutrophils (B), CD11b+ F4/80+ macrophages (C), and inflammatory macrophages (CD11b+ F4/80+ MHC-II+, CD11b+ F4/80+ CD86+, and CD11b+ F4/80+ CD40+, respectively) (D) was assessed. One hundred thousand cells were counted, and the data are expressed as number of positive cells. Data are expressed as means ± standard errors (SE) of the mean and are representative of two independent experiments (*, P < 0.05, and **, P < 0.01).
DISCUSSION
A family of 20 Nsd mutants were first identified in A. nidulans and named based on the fact that they failed to produce cleistothecia, i.e., were never in sexual development (43, 44). Of these mutants, only the NsdC and NsdD mutants have so far been characterized (29, 44, 45). Partly due to the nomenclature used, both the nsdC and nsdD genes are known primarily for their essential role in sexual reproduction (27). However, results from the present study, combined with reevaluation of previous studies of nsdC gene function, reveal that the NsdC transcription factor in particular has much more broad biological functions beyond simply promotion of sexual reproduction, as will now be described.
An indication of one of the additional functions of NsdC came recently from a study aiming to identify new transcription factors involved in the calcium stress response in A. fumigatus (30). Large-scale phenotypic screenings have been widely used to identify new components of signaling pathways and to study drug resistance mechanisms (46–49). A library of null-mutant transcription factors was challenged with high concentrations of CaCl2, and this resulted in the identification of an A. fumigatus ΔnsdC mutant with increased sensitivity to CaCl2 relative to the wild-type parental strain, suggesting a role for NsdC in calcium response. Additional work in the present study demonstrated that complementation with a functional nsdC gene restored the wild-type phenotype. Further phenotyping assays then showed that loss of nsdC led to cyclosporine (a calcineurin-inhibitory drug) resistance. These combined results indicate a first additional important role for NsdC in mediating calcium tolerance. Whether this involves a direct or indirect link to the central calcium-calcineurin-CrzA pathway requires further investigation. Additional experiments are necessary to characterize the interactions among these different TFs and calcineurin. Interestingly, transcriptional profiling using PolII ChIP-Seq showed at least 933 changes in gene expression between the ΔnsdC mutant and the wild type when exposed to calcium stress, indicating a possible wide spectrum of activity for NsdC in calcium response. However, most of these changes are in increased gene expression (805 changes), suggesting that NsdC could act as a repressor.
Given that nsdC was first identified in A. nidulans due to a total lack of sexual development (including failure to produce Hülle cells as well as cleistothecia) even under conditions which favor fruiting body formation by single isolates (29), we then wished to determine whether NsdC had a similar role in sexual development of A. fumigatus. Despite sexual development being demonstrated in A. fumigatus under laboratory conditions (22), most advances in understanding the regulation of the sexual cycle in Aspergillus species have come from studies using the model A. nidulans (27, 44, 50). However, notable differences in the breeding systems of the species (A. fumigatus is heterothallic whereas A. nidulans is homothallic) suggested that findings from A. nidulans might not necessarily be applicable to A. fumigatus. Therefore, both MAT1-1 and MAT1-2 ΔnsdC mutant strains of A. fumigatus were constructed in a high-fertility genetic background to assess the contribution of NsdC in sexual reproduction in this species. It was found that, provided one of the MAT1-1 or MAT1-2 mating partners contained a functional copy of nsdC, the sexual cycle could be completed, leading to cleistothecium and ascospore production, i.e., the absence of NsdC in one mating partner could be complemented by the presence of NsdC produced by the other partner. Indeed, in the original work of Kim et al. (29) it was found that despite the failure to undergo sexual development under selfing conditions, an nsdC deletion mutant of A. nidulans was nevertheless still able to outcross to a compatible auxotrophic strain of A. nidulans with a functional copy of nsdC, mirroring our findings with A. fumigatus. The exact mechanism of complementation is unclear, but it can be speculated that the partner with the functional copy of nsdC might act as the maternal partner, forming ascogonial coils (and later maternal tissues) that could be fertilized by the male ΔnsdC partner, given that nsdC is thought to act at a very early stage of sexual development (27, 29). Alternatively, the partner with the functional copy of nsdC might allow hyphal fusions necessary for sex to occur, with this ability missing from ΔnsdC strains. It has proved difficult to determine the early morphological stages of sexual development in A. fumigatus (K. M. Lord, N. D. Read, and P. S. Dyer, unpublished results), although putative ascogonial coils have been described in A. nidulans (51). However, if both the MAT1-1 and MAT1-2 mating partners of A. fumigatus were of the ΔnsdC genotype, then sexual fertility was totally abolished. This was a significant result, as it demonstrated that NsdC has a second, essential positive regulatory role for sexual development in A. fumigatus, as observed for its counterpart in A. nidulans (29). NsdC has also been shown to have a role in sexual development in A. flavus, where its deletion led to inhibition of formation of sclerotia, these structures being an important first stage in the sexual cycle (27, 52).
These findings also provide a key precedent when evaluating the function of genes in sexual development in the heterothallic A. fumigatus, in that it will be important to disrupt the gene(s) under study in both mating partners to conclusively evaluate gene function. Indeed, it is noted that the function of genes in sexual development is generally performed with homothallic species such as A. nidulans, Sordaria macrospora, and Fusarium graminearum (27, 53, 54) as this removes the time-consuming need to delete a gene(s) in both partners, which is the case for heterothallic species. However, there may be exceptions such as studies of functionality of mating-type genes, as distinct MAT1-1 and MAT1-2 idiomorph genes are found in the different mating partners (20, 25, 55). Also, in contrast to the current results for nsdC, the loss of the nsdD gene (another transcription factor involved in sexual reproduction in the aspergilli) in one mating partner alone was enough to suppress fruit body formation in A. fumigatus (25).
In the original study of nsdC function in A. nidulans, ΔnsdC mutants were also found to be characterized by retarded vegetative growth compared to the wild type, with an approximately 30% reduction in growth rate (29). A very similar result was found in the present study, with the A. fumigatus ΔnsdC strains in both the CEA17 and supermater genetic backgrounds showing a reduction in vegetative growth rate on solid media. However, this growth difference is observed only in minimal medium and not in complete medium, suggesting the ΔnsdC mutant has metabolic deficiencies that are supplemented by the complete medium chemical composition. Taken together, these results confirm a third important role for NsdC in the promotion of vegetative growth. This conclusion is supported in part by the gene expression studies from the present study, involving PolII ChIP-Seq, which showed that NsdC is associated with the regulation of several biological processes under basal growth conditions. It is speculated that the upregulation of some biological functions in the ΔnsdC mutant was perhaps to counterbalance the loss of NsdC. Interestingly, biosynthesis of different aspects of secondary metabolism was up- and downregulated in the ΔnsdC mutant, implying a regulatory function of NsdC in secondary metabolite production as has been observed in Aspergillus flavus (52).
In the original study of nsdC function in A. nidulans, ΔnsdC mutants were furthermore found to be characterized by “hyperactive” asexual sporulation, with earlier-than-normal development of conidia, and conidiation even in liquid media, indicating that NsdC negatively regulates asexual reproduction (29). A very similar result was found in the present study with A. fumigatus, with microscopic examination revealing the production of conidiophores and conidia in the ΔnsdC mutant when grown in liquid MM, conditions under which the parental wild type failed to produce conidia. This activity of NsdC appears to be explained in part as a result of repression of brlA expression. BrlA is a key regulator of asexual sporulation due to its activity in controlling the initiation of conidiophore development (6, 9, 11, 56). It was found that brlA mRNA accumulated in the ΔnsdC strain after 24 h of growth in liquid MM, unlike the parental strain. Higher brlA expression has also been observed in an ΔnsdC mutant of A. flavus (which also exhibited altered conidiophore morphology), and brlA transcripts were found to be expressed earlier (but in similar quantities) in an A. nidulans ΔnsdC mutant (29, 52). Transcriptional profiling also indicated that several genes important for conidiation, such as flbB, flbC, and flbD, have increased expression in the ΔnsdC mutant. Meanwhile, our group and others have previously described a role of the MAP kinase MpkB in the negative regulation of asexual development in both A. fumigatus and A. nidulans, whereby ΔmpkB mutants produced conidia in submerged cultures with a constant accumulation of brlA mRNA (18, 57). In the present study, we observed an accumulation of nsdC mRNA in the ΔmpkB strain of A. fumigatus, indicating negative control of nsdC expression by MpkB. Further investigation is required in order to establish if MpkB also influences sexual development in A. fumigatus, as already described for A. nidulans (28). Taken together, these results confirm a fourth important role for NsdC in the negative regulation of asexual reproduction by transcriptional repression of brlA, subject to control by MpkB.
Cell wall composition and structure vary substantially with cell cycle progression and in response to environmental changes (58). Very little is known about A. fumigatus TFs regulating cell wall biosynthesis and/or remodeling. A. fumigatus TFs such as RlmA and calcium-calcineurin-dependent TFs such as CrzA and ZipD have been characterized in more detail as regulating several genes involved in the cell wall metabolism (30, 49, 59–66). In the present study, deletion of the nsdC gene led to greater sensitivity to cell wall-damaging agents, a reorganization of cell wall structure, decreased and increased cell wall sugar content, and increased cell wall width. The A. fumigatus cell wall is composed of more than 90% polysaccharides, with the core skeleton composed by a branched β-1,3-glucan linked to chitin, galactomannan, and β-1,3-β-1,4-glucans; α-1,3-glucan and mannans act as a cement, filling the pores between fibrillar polysaccharides (67). We have observed no differences in total carbohydrates isolated from the cell walls of the wild-type, ΔnsdC, and complemented strains. However, there are increased concentrations of mannose and galactose and decreased concentrations of glucosamine, glucose, and NAG present in the ΔnsdC cell walls compared to the wild-type and complemented strains. It proved difficult to establish a direct relationship between the organization and composition of the fungal cell wall that could directly explain the observed increased thickness of ΔnsdC cell walls. It is possible that the combination of the increased and decreased sugar concentrations could impact the ΔnsdC cell wall organization, increasing the cell wall width. Transcriptional profiling also showed that several genes involved in the cell wall biosynthesis and remodeling were dysregulated in the ΔnsdC mutant. Cell wall thickening has also been linked to defects in sphingolipid metabolism in A. nidulans and yeasts (68), but alteration of cell membrane components was not assessed in this work. Interestingly, transcriptional profiling found up- and downregulated genes involved in lipid, fatty acid, and isoprenoid metabolism, which could help to explain this striking phenotype. In contrast, loss of nsdD in A. fumigatus conferred resistance toward certain cell wall stressors (25). Taken as a whole, these results demonstrate a fifth important role for NsdC in response to cell wall stress and correct cell wall organization. More studies are necessary to understand how NsdC interacts with RlmA, CrzA, and ZipD; is regulated by calcineurin; and populates the promoter regions of the genes encoding proteins important for the cell wall integrity pathway.
Cell wall organization impacts directly virulence and host immune recognition in A. fumigatus (69). Accordingly, leukopenic mice inoculated with ΔnsdC conidia showed a reduction in mortality, fungal burden, and inflammation compared to inoculation with the wild-type parental strain, possibly also linked to the lower growth rate of the ΔnsdC strain. Despite the reduced virulence, ΔnsdC conidia were less well recognized by macrophages than wild-type conidia, probably because of the increased thickness of the cell wall in this mutant which possibly masks β-1,3-glucan (which is already reduced in the mutant) and other cell wall components responsible for triggering immune response in the host (70). As a result, ΔnsdC conidia were also less efficiently eliminated. This impaired interaction between macrophages and ΔnsdC conidia likely resulted in fewer inflammatory cytokines and consequently a diminished influx of leukocytes to the site of inoculation as further confirmed by results from the immunocompetent model of pulmonary acute aspergillosis included in the present study. Corroborating the lower phagocytosis index, the killing rates may be influenced not just because the conidia were not killed but also because the conidium ingestion was affected. However, a compromised phagocytosis associated with an efficient macrophage activity would affect the recruitment of immune cells as shown in Fig. 6A. The impaired interaction between alveolar macrophages and the nsdC mutant strain likely resulted in reduced inflammatory signals and consequently less influx of leucocytes to the site of infection as we showed in Fig. 6. In addition, the diminished expression of activation markers (Fig. 6D) clearly showed that mechanisms of macrophage activation were impaired against the nsdC mutant strain. We believe that the cell wall organization impacts directly virulence and host immune recognition by innate immune cells during A. fumigatus infection. These findings were previously described and revised by reference 69. These results demonstrate an extra sixth role for NsdC in virulence and host immune recognition.
In summary, in addition to its known essential roles in sexual reproduction and control of growth rate and asexual reproduction, we have shown in the present study of A. fumigatus that the NsdC transcription factor has additional previously unrecognized biological functions including calcium tolerance, cell wall stress response, and correct cell wall organization and functions in virulence and host immune recognition. Indeed, the fact that NsdC is needed for correct vegetative growth and that gene deletion resulted in changes in expression of over 620 genes under basal growth conditions suggests that promotion of sexual reproduction is perhaps not the principal role of NsdC (despite the gene epithet) and that instead this transcription factor mediates the activity of a wide variety of key signaling and metabolic pathways. In contrast, deletion of many other genes in A. nidulans with a principal role in sexual reproduction has little impact on vegetative growth (27). In addition, whereas overexpression of the key sex-related genes veA and nsdD resulted in induction of sex in submerged cultures of A. nidulans, no such effect was seen with overexpression of nsdC, again consistent with a primary role other than sexual reproduction (29). The multifunctionality of NsdC has been proposed to be linked to the transcription of two different mRNA forms from the nsdC gene in A. nidulans, with a 3.0-kb form staying relatively constant during the life cycle whereas a shorter, 2.6-kb form accumulated differentially especially during sterigma and sexual development (29). Further research is now warranted into how gene expression is controlled by the NsdC transcription factor.
MATERIALS AND METHODS
Strains and media.
All strains used in this study are listed in either Table S1 or Table S5 at https://doi.org/10.6084/m9.figshare.12931754.v3. Strains for genetic manipulation were grown at 37°C in either complete medium (YG: 2% [wt/vol] glucose, 0.5% [wt/vol] yeast extract, trace elements) or minimal medium (MM: 1% [wt/vol] glucose, original high-nitrate salts, trace elements, pH 6.5). Solid YG and MM were the same as described above except that 2% (wt/vol) agar was added. Trace elements, vitamins, and nitrate salts compositions were as described previously (71). When required, MM was supplemented at stated concentrations with calcium chloride (CaCl2), cyclosporine, sorbitol, nikkomycin Z, Congo red (CR), calcofluor white (CFW), and caspofungin. Strains for sexual reproductive purposes were grown at 28°C on Aspergillus complete medium for production of conidia for mating inoculum (20). For characterization of phenotype, plates were inoculated with 104 spores per strain and left to grow for 120 h at 37°C.
Construction of A. fumigatus mutants for ΔnsdC growth assays and NsdC-GFP localization.
To generate the NsdC-GFP::pyrG fusion fragment, a 3-kb portion of DNA consisting of the nsdC open reading frame (ORF) and 5′ untranslated region (UTR), along with a 1-kb segment of DNA consisting of the 3′ UTR flanking region, was amplified with primers nsdC pRS426 5fw/nsdC orf LINKER GFP rv and nsdC 3utr pyrG 3fw/nsdC pRS426 3rv, respectively, from CEA17 genomic DNA (gDNA). The 3.3-kb linker-GFP-trpC-pyrG fusion was amplified with primers OZG916 and OZG964 from the pOB435 plasmid. The cassette was generated by transforming each fragment along with the plasmid pRS426 cut with BamHI/EcoRI into the Saccharomyces cerevisiae strain. This cassette was then transformed into the CEA17 strain, and verification of NsdC tagging was confirmed via PCR. Figures S4 and S5 at https://doi.org/10.6084/m9.figshare.12931754.v3 show the confirmatory PCR and Southern blots for the deletion, complementation, and GFP fusion strains. All primers used above are described in Table S6 at https://doi.org/10.6084/m9.figshare.12931754.v3.
Microscopy.
Conidia from the NsdC-GFP strain were cultivated on coverslips in 4 ml of MM for 16 h at 30°C before treatment with 200 mM CaCl2 for 15 min, 0.25 μg/ml cyclosporine for 30 min, or 0.25 μg/ml cyclosporine for 30 min and 200 mM CaCl2 for 15 min. For colocalization experiments, germlings were costained with Hoechst stain (12 μg/ml) (Life Technologies, Inc.) for 3 min. Coverslips were inspected on an Observer Z1 fluorescence microscope (Carl Zeiss) using the 100× objective oil immersion lens (for GFP, filter set 38 HE, excitation wavelength of 450 nm to 490 nm, and emission wavelength of 525 nm to 550 nm; for Hoechst stain, filter set 49 HE, excitation wavelength of 365 nm, and emission wavelength of 420 nm to 470 nm). Differential interference contrast (DIC) images and fluorescent images were captured with an AxioCam camera (Carl Zeiss, Inc.) and processed using the AxioVision software (version 4.8).
Conidium count.
Freshly harvested conidia (1 × 104) of the wild-type and ΔnsdC and ΔnsdC::nsdC+ mutant strains were inoculated onto solid MM at 37°C for 5 days. After this period of growth, four circular sections of the same size were taken from each plate (approximately 1 cm in diameter each) and placed in a Falcon tube containing 10 ml of a 0.01% Tween solution. Conidia were counted after intensive vortexing using a Neubauer chamber. The assay was performed in triplicate.
Generation of ΔnsdC mutants in a high-fertility MAT1-1 background.
The highly fertile A. fumigatus MAT1-1 strain 47-51 (synonym AfIR974 [22]) was used as a recipient strain to generate an ΔnsdC mutant. The nsdC deletion construct was amplified by Phusion polymerase (Thermo) from plasmid pRS426 with primers Af_nsdCSma_up_f and Af_nsdCSma_do_r (see Table S6 at https://doi.org/10.6084/m9.figshare.12931754.v3), resulting in a PCR product with flanking SmaI restriction sites. The fragment was cloned into the pJET1.2 PCR cloning vector (Thermo) and amplified in Escherichia coli DH5α cells. Plasmid DNA was isolated by using the NucleoSpin plasmid isolation kit (Macherey-Nagel) and restricted with SmaI (FastDigest; Thermo) to release the nsdC deletion cassette from the plasmid backbone. The 4,345-bp deletion construct, including the pyrithiamine resistance cassette ptrA flanked by the upstream and downstream noncoding regions of the nsdC gene, was purified from a 1% agarose gel using the GeneJET gel extraction kit (Thermo), and about 2.5 μg of DNA was used for a polyethylene glycol (PEG)-mediated transformation of A. fumigatus strain 47-51 as described previously (72), with the exception that a mixture of 1.2 g VinoTaste Pro (Novozymes), 0.1 g lysing enzymes from Trichoderma harzianum (Sigma), and 0.1 g Yatalase (TaKaRa/Clontech) was used for the generation of protoplasts. Transformants were selected by the presence of 0.1 μg/ml pyrithiamine (Sigma) in the regeneration medium. Transformants were subsequently analyzed for gene deletion by Southern blot analysis. A digoxigenin (DIG)-labeled probe against the nsdC upstream region was amplified by Taq polymerase (New England Biolabs) with primers Af_nsdCSma_up_f and nsdCAf_up_r (see Table S6 at https://doi.org/10.6084/m9.figshare.12931754.v3) and a nucleotide mix containing DIG-11-dUTP. Genomic DNA of wild type and transformants was restricted with EcoRI, separated on a 0.8% agarose gel, blotted on a nylon membrane, and hybridized with the digoxigenin-labeled probe. Bands were visualized by using antidigoxigenin Fab fragments linked to alkaline phosphatase and developed by using the chemiluminescent substrate CDP-Star (Sigma). Transformants showing a single band and a shift of the wild-type signal from 5.3 kb down to 2 kb were used in subsequent experiments.
Generation of ΔnsdC mutants with a MAT1-2 background.
To test the fertility of the ΔnsdC mutants in the MAT1-1 background of the high-fertility strain 47-51 and to generate nsdC mutants with a MAT1-2 background, sexual crosses were set up with high- to medium-fertility MAT1-2 strains of A. fumigatus, namely, strains 47-55 (synonym AfIR964 [22]) and 47-107 (S. S. Swilaiman, G. Szakacs, and P. S. Dyer, unpublished data) (see Tables S1 and S2 at https://doi.org/10.6084/m9.figshare.12931754.v3). Crosses were set up on oatmeal agar at 30°C as described by Ashton and Dyer (24) with four replicate 9-cm petri plates per cross, together with control crosses with the 47-51 parent. After 3 to 4 months of incubation, total numbers of cleistothecia per plate were counted using a dissecting microscope and “hoovering” conidia from plates to ensure that any cleistothecia were visible (23, 24). Cleistothecia were collected, and ascospores were released into 100 μl sterile 0.05% Tween 80. Ascospores were then heated for 1 h at 70°C to inactivate remaining conidia and vegetative hyphae and to simultaneously induce germination of ascospores (22, 24). Ascospores were plated on Aspergillus minimal medium with 50 mM glucose as a carbon source and 10 mM glutamine as a nitrogen source (73) in the presence or absence of 0.1 μg/ml of pyrithiamine, given that pyrithiamine resistance was indicative of deletion of the nsdC gene. The mating type of individual colonies was subsequently determined by multiplex PCR as previously described (20).
Fertility analyses of ΔnsdC mutants with either MAT1-1 or MAT1-2 mating-type background.
Sexual offspring of MAT1-1 or MAT1-2 mating type which were of the ΔnsdC genotype were analyzed for sexual fertility in crossing experiments with either wild type or ΔnsdC mutants of the opposite mating type. This included the use of an additional high-fertility MAT1-1 isolate, 47-267 (see Table S1 at https://doi.org/10.6084/m9.figshare.12931754.v3), for testing of the fertility of MAT1-2 isolates. Four replicate 9-cm petri plates were set up for each cross. The level of fertility was evaluated by counting total numbers of cleistothecia per crossing plate as described above. Resultant data were analyzed using Prism 8.0 by one-way analysis of variance (ANOVA) and nested one-way ANOVA as appropriate.
RNA extraction and gene expression analysis.
Strains were grown from 1 × 107 conidia in MM for 24 h (wild-type, ΔnsdC, ΔnsdC::nsdC+, and ΔmpkB strains) or 48 h (wild-type and ΔmpkB strains) at 37°C. Mycelia were ground to a fine powder in liquid N2, and total RNA was extracted with TRIzol reagent (Thermo Scientific) according to the manufacturer’s protocol. DNA was digested with Turbo DNase I (Ambion Thermo Scientific) according to the manufacturer’s instructions. Two micrograms of total RNA per sample was reverse transcribed with the High-Capacity cDNA reverse transcription kit (Thermo Scientific) using oligo dTV and random primer blend, according to manufacturer’s instructions. qRT-PCRs were run in a StepOne Plus real-time PCR system (Thermo Scientific) using a Power Sybr green PCR master mix (Thermo Scientific). Three independent biological replicates were used, and the mRNA quantity relative fold change was calculated using standard curves (74). All values were normalized to the expression of the A. fumigatus tubA gene. Primers are described in Table S6 at https://doi.org/10.6084/m9.figshare.12931754.v3.
PolII chromatin immunoprecipitation coupled to DNA sequencing (PolII ChIP-Seq).
Conidia (1 × 107) of the CEA17 and ΔnsdC strains were grown in liquid MM for 16 h at 37°C under shaking conditions. Mycelia and chromatin were prepared as previously described (75). Briefly, formaldehyde was added to the culture to a final concentration of 1% and incubated with gentle rocking for 20 min at room temperature for DNA cross-linking. After, glycine was added to a final concentration of 1 M and the culture was incubated for another 10 min to stop the reaction. Cells were filtered using Miracloth, washed twice with 100 ml of cold water, and rapidly frozen in liquid nitrogen. Thirty milligrams of the frozen mycelia were freeze-dried for at least 2 h and lysed in 800 μl of FA lysis buffer for 3 min using a Bullet blender (Next Advance). Lysis was repeated six times with a 3-min incubation on ice in between. The lysed mycelium was pelleted by centrifugation at 14,000 rpm for 15 min at 4°C and subsequently resuspended in 500 μl of FA lysis buffer and sonicated using the Qsonica Q800R at 100% amplitude with 10-s ON and 15-s OFF cycles for a total sonication time of 30 min. The arising chromatin solution was recovered by centrifugation at 14,000 rpm for 30 min at 4°C and stored at −80°C until use. Chromatin size (∼100 to 300 bp) and quality were checked on a 2% agarose gel. PolII immunoprecipitation was performed by mixing 50 μl of chromatin extract with 450 μl of FA lysis buffer and 2 μl of anti-RNA polymerase II subunit antibodies (clone 3E8; Millipore) for 1.5 h, followed by incubating the mixture with ∼15 μl packed protein A Sepharose (GE Healthcare) for another 1.5 h at room temperature on an end-to-end rotator. Subsequently, protein A Sepharose matrix was transferred to a Corning Costar SpinX centrifuge tube filter and washed. Immunoprecipitated chromatin DNA was de-cross-linked at 65°C overnight and then was purified using a Qiagen PCR cleanup purification column. Library preparation was carried out using an NEBNext Ultra II library prep kit (Illumina; catalog no. E7645L) according to the manufacturer’s protocol and barcoded adaptors as described in the work of Wong et al. (76). Libraries were checked and quantified using a DNA High Sensitivity Bioanalyzer assay (Agilent; catalog no. XF06BK50), mixed in equal molar ratio, and sequenced using the Illumina HiSeq2500 platform at the Genomics and Single Cells Analysis Core facility at the University of Macau.
Raw sequencing reads were mapped to the A. fumigatus strain Af293 reference genome (AspGD version s03-m05-r06) using Bowtie2 (v2.3.5) (77). PolII binding levels were measured by counting number of reads over coding regions and normalizing as FPKM (fragments per kilobase per million) scores. For ChIP-Seq signal data visualization purpose, aligned reads were extended to 200 bp using the ‘macs2 pileup’ command and scaled to 1 million mapped reads using the ‘macs2 bdgopt’ command from the MACS2 (v2.1.1) tool (78). UCSC Kent Utils programs (79) ‘bedSort’ and ‘bedGraphToBigWig’ were used to generate BigWig files, which were visualized in Integrative Genomics Viewer (v2.5.3) (80).
Cell wall polysaccharide extraction and sugar quantification.
Fungal cell wall polysaccharides were extracted from 100 mg dry-frozen biomass as described previously using TCA (trichloroacetic acid) hydrolysis (81). Total carbohydrates were estimated using the phenol sulfuric method as described by Masuko et al. (82). Released sugars from hydrolysis were subsequently analyzed by high-performance liquid chromatography (HPLC) using a YoungLin YL9100 series system (YoungLin, Anyang, South Korea) equipped with a YL9170 series refractive index (RI) detector at 40°C. Samples were loaded in a Rezex ROA (Phenomenex, USA) column (300 × 7.8 mm) at 85°C and eluted with 0.05 M sulfuric acid at a flow rate of 0.5 ml/min. All sugar concentrations were expressed in millimolar (mM) using a correspondent standard curve.
Staining for dectin-1, chitin, and other cell surface carbohydrates.
Cell wall surface polysaccharide staining was performed as described previously (83, 84). Briefly, strains were grown from 2.5 × 103 spores in 200 μl of MM for 16 h at 37°C before the culture medium was removed and germlings were UV irradiated (600,000 μJ). Hyphal germlings were subsequently washed with 1× phosphate-buffered saline (PBS), and 200 μl of a blocking solution (2% [wt/vol] goat serum, 1% [wt/vol] bovine serum albumin [BSA], 0.1% [vol/vol] Triton X-100, 0.05% [vol/vol] Tween 20, 0.05% [vol/vol] sodium azide, and 0.01 M PBS) was added. Samples were incubated for 30 min at room temperature (RT). For dectin staining, 0.2 μg/ml of Fc-h-dectin-hFc was added to the UV-irradiated germlings and incubated for 1 h at RT, followed by the addition of 1:1,000 DyLight 594-conjugated, goat anti-human IgG1 for 1 h at RT. Germlings were washed with PBS, and fluorescence was read at 587-nm excitation and 615-nm emission. For chitin staining, 200 μl of a PBS solution with 10 μg/ml of calcofluor white (CFW) was added to the UV-irradiated germlings, which were incubated for 5 min at RT and washed three times with PBS before fluorescence was read at 380-nm excitation and 450-nm emission. For galactosaminogalactan (GAG), GlcN (glucosamine), and mannose staining, 200 μl of PBS supplemented with 0.1 mg/ml of either soybean agglutinin-fluorescein isothiocyanate (SBA-FITC) (Glycine max soybean lectin SBA-FITC; Bioworld; catalog no. 21761024-2), wheat germ agglutinin (WGA) (lectin-FITC L4895; Sigma), or concanavalin A (ConA; C7642; Sigma) was added to the UV-irradiated germlings for 1 h at RT. Germlings were washed with PBS, and fluorescence was read at 492-nm excitation and 517-nm emission. All experiments were performed using 12 repetitions, and fluorescence was read in a microtiter plate reader (SpectraMax i3; Molecular Devices).
Transmission electron microscopy (TEM) analysis of cell wall.
Strains were grown statically from 1 × 107 conidia at 37°C in MM for 24 h. Mycelia were harvested and immediately fixed in 0.1 M sodium phosphate buffer (pH 7.4) containing 2.5% (vol/vol) glutaraldehyde and 2% (wt/vol) paraformaldehyde for 24 h at 4°C. Samples were encapsulated in agar (2%, wt/vol) and subjected to fixation (1% OsO4), contrasting (1% uranyl acetate), ethanol dehydration, and a two-step infiltration process with Spurr resin (Electron Microscopy Sciences) of 16 h and 3 h at RT. Additional infiltration was provided under vacuum at RT before embedment in BEEM capsules (Electron Microscopy Sciences) and polymerization at 60°C for 72 h. Semithin (0.5-μm) survey sections were stained with toluidine blue to identify the areas of best cell density. Ultrathin sections (60 nm) were prepared and stained again with uranyl acetate (1%) and lead citrate (2%). Transmission electron microscopy (TEM) images were obtained using a Philips CM-120 electron microscope at an acceleration voltage of 120 kV using a MegaView3 camera and iTEM 5.0 software (Olympus Soft Imaging Solutions GmbH). Cell wall thicknesses of 100 sections of different germlings were measured at ×23,500 magnification, and images were analyzed with the ImageJ software (85). Statistical differences were evaluated by using one-way analysis of variance (ANOVA) and Tukey’s post hoc test.
Ethics statement.
Animal experiments were performed in strict accordance with Brazilian Federal Law 11794, establishing procedures for the scientific use of animals in strict accordance with the principles outlined by the Brazilian College of Animal Experimentation (CONCEA), and the state law establishing the animal protection code of the State of São Paulo. All protocols adopted in this study were approved by the local ethics committee for animal experiments from the University of São Paulo, Campus of Ribeirão Preto (permit number 08.1.1277.53.6; Studies on the interaction of Aspergillus fumigatus with animals). All efforts were made to minimize suffering. All stressed animals were sacrificed by cervical dislocation after intraperitoneal (i.p.) injection of ketamine and xylazine.
Murine model of pulmonary aspergillosis, lung histopathology, and fungal burden.
Outbred female mice (BALB/c strain; body weight, 20 to 22 g) were housed in vented cages containing five animals. Mice were immunosuppressed with cyclophosphamide (150 mg per kg of body weight), which was administered intraperitoneally on days −4, −1, and 2 prior to and after infection. Hydrocortisonacetate (200 mg/kg body weight) was injected subcutaneously on day −3. A. fumigatus strains were grown on YAG for 2 days prior to infection. Fresh conidia were harvested in PBS and filtered through Miracloth (Calbiochem). Conidial suspensions were spun for 5 min at 3,000 × g, washed three times with PBS, counted using a hemocytometer, and resuspended at a concentration of 5.0 × 106 conidia/ml. The viability of the administered inoculum was determined by incubating a serial dilution of the conidia on YAG medium, at 37°C. Mice were anesthetized by halothane inhalation and infected by intranasal instillation of 1.0 × 105 conidia in 20 ml of PBS. As a negative control, a group of five mice received PBS only. Mice were weighed every 24 h from the day of infection and visually inspected twice daily. The statistical significance of the comparative survival values was calculated using log rank analysis and the Prism statistical analysis package (GraphPad Software Inc.).
To investigate fungal burden in murine lungs, mice were immunosuppressed with cyclophosphamide (150 mg/kg of body weight), which was administered intraperitoneally on days −4 and −1, while hydrocortisonacetate was injected subcutaneously (200 mg/kg) on day −3. Five mice per group were intranasally inoculated with 1 × 106 conidia/20 ml of suspension. A higher inoculum, in comparison to the survival experiments, was used to increase fungal DNA detection. Animals were sacrificed 72 h postinfection, and the lungs were harvested and immediately frozen in liquid nitrogen. Samples were homogenized by vortexing with glass beads for 10 min, and DNA was extracted via the phenol-chloroform method. DNA quantity and quality were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific). At least 500 mg of total DNA from each sample was used for quantitative real-time PCRs. Specific primers were used to amplify the 18S rRNA region of A. fumigatus and an intronic region of mouse GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (see Table S6 at https://doi.org/10.6084/m9.figshare.12931754.v3). Six-point standard curves were calculated using serial dilutions of gDNA from all the A. fumigatus strains used and the uninfected mouse lung. Fungal and mouse DNA quantities were obtained from the threshold cycle (CT) values from an appropriate standard curve (74). Fungal burden was determined as the ratio between picograms of fungal DNA and micrograms of mouse DNA.
For histopathology studies, the animals were also sacrificed 72 h postinfection and lungs were removed and fixed for 24 h in 3.7% formaldehyde-PBS. Samples were washed several times in 70% alcohol before dehydration in a series of alcohol solutions of increasing concentrations. Finally, samples were diaphanized in xylol and embedded in paraffin. For each sample, sequential 5-mm-thick sections were collected on glass slides and stained with Grocott’s methenamine silver (GMS) or hematoxylin and eosin (HE) stain following standard protocols. Briefly, sections were deparaffinized, oxidized with 4% chromic acid, stained with methenamine silver solution, and counterstained with picric acid. For HE staining, sections were deparaffinized and stained first with hematoxylin and then with eosin. All stained slides were immediately washed, preserved with mounting medium, and sealed with a coverslip. Microscopic analyses were done using an Axioplan 2 imaging microscope (Zeiss) at the stated magnifications under bright-field conditions.
A. fumigatus infection in immunocompetent mice.
Eight- to 12-week-old male C57BL/6 WT mice were obtained from the specific-pathogen-free Isogenic Breeding Unit of the Department of Immunology, Institute of Biomedical Sciences, University of São Paulo. Mice were anesthetized and submitted to intratracheal (i.t.) infection as previously described (86) and further adapted to the A. fumigatus infection model (30, 87). Briefly, after intraperitoneal (i.p.) injection of ketamine and xylazine, animals were inoculated with 5 × 107 CEA17 wild-type, ΔnsdC, or ΔnsdC::nsdC+ conidia, contained in 75 μl of PBS, by surgical i.t. inoculation, which allowed dispensing of the fungal cells directly into the lungs.
Assessment of leukocyte subpopulations and flow cytometric analysis.
Lungs from A. fumigatus-inoculated mice were collected after 3 days of exposure to the microbe. To assess the leukocyte subpopulations, the lungs were removed and digested enzymatically for 40 min with collagenase (2 mg/ml) in RPMI culture medium (Sigma). Total lung leukocyte numbers were assessed with trypan blue, and viability was always >95%. For cell surface staining, leukocytes were washed and suspended at 1 × 106 cells/ml in staining buffer (PBS, 2% fetal calf serum, and 0.1% NaN3). Fc receptors were blocked by the addition of unlabeled anti-CD16/32 (eBioscience). Leukocytes were then stained in the dark for 25 min at 4°C with the optimal dilution of each monoclonal antibody. The following were added to total leukocytes, neutrophils, and macrophages: anti-CD45, CD11b, Ly6G, Ly6C, F4/80, CD40, MHC-II, and CD86 (eBioscience or BioLegend). Cells were washed twice with staining buffer, before being fixed with 2% paraformaldehyde (PFA; Sigma). A minimum of 100,000 events was acquired on a FACSLyrics flow cytometer (BD Biosciences) using the FACSDiva software (BD Biosciences). Total leukocytes, macrophages, and neutrophils were gated as judged from forward and side light scatter. The cell surface expression of leukocyte markers was analyzed using the FlowJo software (Tree Star).
Bone marrow-derived macrophage (BMDM) preparation, phagocytosis, and killing assay.
The BMDM preparation was performed according to the work of Marim et al. (88) with some modifications. BMDMs were prepared from C57BL/6 adult mouse femurs and tibias. These cells were cultured for 6 days in RPMI 1640 medium supplemented with 20% fetal cow serum (FCS) and 30% L-929 cell-conditioned medium. Nonadherent cells were removed, and the adherent cells (majority macrophages) were removed and washed twice with cold PBS. Cell concentration was determined using a Neubauer chamber. The phagocytic assay was performed as previously described (89) with slight modifications. In 24-well microplates at 37°C with 5% CO2, 1 ml of RPMI-FCS containing 1 × 105 conidia (1:5 macrophage/conidium ratio) was added and incubated for 90 min. The supernatant was removed, and 0.5 ml of 3.7% formaldehyde-PBS was added. The samples were washed with ultrapure water and incubated for 20 min with 495 μl of water and 5 μl of CFW (10 mg/ml). Samples were washed and mounted on slides with 50% glycerol. Images were then acquired on an Eclipse E800 fluorescence microscope (Nikon Instruments), and the phagocytosis index was calculated counting at least 100 conidia per sample. The experiments were repeated in triplicate. To assess conidial killing, the phagocytic cells were obtained as described above. Subsequently, 1 × 105 conidia (1:5 macrophage/conidium ratio) were incubated at 37°C with 5% CO2 for 4 h. As positive control, conidia without BMDMs were used. The 24-well microplates were centrifuged for 10 min at 3,500 rpm, and the culture supernatant was removed. Then, 100 μl of 1% Triton X-100 was added. After 10 min at room temperature, samples were removed from the microplate, washed three times with sterile distilled water, and serially diluted in PBS. The dilutions were plated and incubated at 37°C for 48 h. The percentage of conidial killing was calculated by the measurement of CFU numbers carried out after macrophage lysis, comparing CFU numbers from samples incubated with macrophages to CFU numbers from those incubated without macrophages. The experiments were repeated three times, each performed in triplicate.
Data availability.
The transcriptional profiling for ΔnsdC was investigated using ChIP of RNA polymerase II coupled to DNA sequencing. Short reads were deposited in the NCBI, under GEO accession number GSE148557.
ACKNOWLEDGMENTS
We thank São Paulo Research Foundation (FAPESP) grant numbers 2016/07870-9 (G.H.G.), 2018/00715-3 (C.V.), 2016/12948-7 (P.A.D.C.), 2016/22062-6 (J.C.), 2016/21392-2 (L.P.S.), 2019/10097-8 (F.V.L.), and 2017/07536-4 (A.C.C.) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (G.H.G.), both from Brazil, for financial support. K.H.W. also acknowledges the support from the Research Services and Knowledge Transfer Office (MYRG2018-00017-FHS and MYRG2019-00099-FHS) of the University of Macau and the Collaborative Research Fund Equipment Grant (C5012-15E) from the Research Grant Council, Hong Kong Government. P.S.D. thanks the Natural Environment Research Council (UK) for support (NE/P000916/1).
Footnotes
Citation Alves de Castro P, Valero C, Chiaratto J, Colabardini AC, Pardeshi L, Pereira Silva L, Almeida F, Campos Rocha M, Nascimento Silva R, Malavazi I, Du W, Dyer PS, Brock M, Vieira Loures F, Wong KH, Goldman GH. 2021. Novel biological functions of the NsdC transcription factor in Aspergillus fumigatus. mBio 12:e03102-20. https://doi.org/10.1128/mBio.03102-20.
REFERENCES
- 1.Kontoyiannis DP, Bodey GP. 2002. Invasive aspergillosis in 2002: an update. Eur J Clin Microbiol Infect Dis 21:161–172. doi: 10.1007/s10096-002-0699-z. [DOI] [PubMed] [Google Scholar]
- 2.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 3.Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi 3:57. doi: 10.3390/jof3040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregg KS, Kauffman CA. 2015. Invasive aspergillosis: epidemiology, clinical aspects, and treatment. Semin Respir Crit Care Med 36:662–672. doi: 10.1055/s-0035-1562893. [DOI] [PubMed] [Google Scholar]
- 5.Park HS, Yu JH. 2016. Developmental regulators in Aspergillus fumigatus. J Microbiol 54:223–231. doi: 10.1007/s12275-016-5619-5. [DOI] [PubMed] [Google Scholar]
- 6.Ojeda-López M, Chen W, Eagle CE, Gutiérrez G, Jia WL, Swilaiman SS, Huang Z, Park HS, Yu JH, Cánovas D, Dyer PS. 2018. Evolution of asexual and sexual reproduction in the aspergilli. Stud Mycol 91:37–59. doi: 10.1016/j.simyco.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiser DM, Klich MA, Frisvad JC, Peterson SW, Varga J, Samson RA. 2007. The current status of species recognition and identification in Aspergillus. Stud Mycol 59:1–10. doi: 10.3114/sim.2007.59.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyer PS, O’Gorman CM. 2011. A fungal sexual revolution: Aspergillus and Penicillium show the way. Curr Opin Microbiol 14:649–654. doi: 10.1016/j.mib.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Adams TH, Boylan MT, Timberlake WE. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353–362. doi: 10.1016/0092-8674(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 10.Andrianopoulos A, Timberlake WE. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol Cell Biol 14:2503–2515. doi: 10.1128/mcb.14.4.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mah JH, Yu JH. 2006. Upstream and downstream regulation of asexual development in Aspergillus fumigatus. Eukaryot Cell 5:1585–1595. doi: 10.1128/EC.00192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao L, Yu JH. 2011. AbaA and WetA govern distinct stages of Aspergillus fumigatus development. Microbiology (Reading) 157:313–326. doi: 10.1099/mic.0.044271-0. [DOI] [PubMed] [Google Scholar]
- 13.Alkhayyat F, Chang Kim S, Yu JH. 2015. Genetic control of asexual development in Aspergillus fumigatus. Adv Appl Microbiol 90:93–107. doi: 10.1016/bs.aambs.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Park HS, Bayram O, Braus GH, Kim SC, Yu JH. 2012. Characterization of the velvet regulators in Aspergillus fumigatus. Mol Microbiol 86:937–953. doi: 10.1111/mmi.12032. [DOI] [PubMed] [Google Scholar]
- 15.Xiao P, Shin KS, Wang T, Yu JH. 2010. Aspergillus fumigatus flbB encodes two basic leucine zipper domain (bZIP) proteins required for proper asexual development and gliotoxin production. Eukaryot Cell 9:1711–1723. doi: 10.1128/EC.00198-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin KS, Kwon NJ, Yu JH. 2009. Gbetagamma-mediated growth and developmental control in Aspergillus fumigatus. Curr Genet 55:631–641. doi: 10.1007/s00294-009-0276-4. [DOI] [PubMed] [Google Scholar]
- 17.Kwon NJ, Park HS, Jung S, Kim SC, Yu JH. 2012. The putative guanine nucleotide exchange factor RicA mediates upstream signaling for growth and development in Aspergillus. Eukaryot Cell 11:1399–1412. doi: 10.1128/EC.00255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manfiolli AO, Siqueira FS, Dos Reis TF, Van Dijck P, Schrevens S, Hoefgen S, Föge M, Straßburger M, de Assis LJ, Heinekamp T, Rocha MC, Janevska S, Brakhage AA, Malavazi I, Goldman GH, Valiante V. 2019. Mitogen-activated protein kinase cross-talk interaction modulates the production of melanins in Aspergillus fumigatus. mBio 10:e00215-19. doi: 10.1128/mBio.00215-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varga J, Toth B. 2003. Genetic variability and reproductive mode of Aspergillus fumigatus. Infect Genet Evol 3:3–17. doi: 10.1016/s1567-1348(02)00156-9. [DOI] [PubMed] [Google Scholar]
- 20.Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, Debeaupuis JP, Latgé JP, Denning DW, Dyer PS. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol 15:1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 21.Pringle A, Baker DM, Platt JL, Wares JP, Latge JP, Taylor JW. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886–1899. doi: 10.1111/j.0014-3820.2005.tb01059.x. [DOI] [PubMed] [Google Scholar]
- 22.O’Gorman CM, Fuller H, Dyer PS. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- 23.Sugui JA, Losada L, Wang W, Varga J, Ngamskulrungroj P, Abu-Asab M, Chang YC, O’Gorman CM, Wickes BL, Nierman WC, Dyer PS, Kwon-Chung KJ. 2011. Identification and characterization of an Aspergillus fumigatus “supermater” pair. mBio 2:e00234-11. doi: 10.1128/mBio.00234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashton GD, Dyer PS. 2019. Culturing and mating of Aspergillus fumigatus. Curr Protoc Microbiol 54:e87. doi: 10.1002/cpmc.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szewczyk E, Krappmann S. 2010. Conserved regulators of mating are essential for Aspergillus fumigatus cleistothecium formation. Eukaryot Cell 9:774–783. doi: 10.1128/EC.00375-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dyer PS, Kuck U. 2017. Sex and the imperfect fungi. Microbiol Spectr 5:FUNK-0043-2017. doi: 10.1128/microbiolspec.FUNK-0043-2017. [DOI] [PubMed] [Google Scholar]
- 27.Dyer PS, O’Gorman CM. 2012. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev 36:165–192. doi: 10.1111/j.1574-6976.2011.00308.x. [DOI] [PubMed] [Google Scholar]
- 28.Paoletti M, Seymour FA, Alcocer MJC, Kaur N, Calvo AM, Archer DB, Dyer PS. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol 17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Kim HR, Chae KS, Han KH, Han DM. 2009. The nsdC gene encoding a putative C2H2-type transcription factor is a key activator of sexual development in Aspergillus nidulans. Genetics 182:771–783. doi: 10.1534/genetics.109.101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Castro PA, Colabardini AC, Manfiolli AO, Chiaratto J, Silva LP, Mattos EC, Palmisano G, Almeida F, Persinoti GF, Ries LNA, Mellado L, Rocha MC, Bromley M, Silva RN, de Souza GS, Loures FV, Malavazi I, Brown NA, Goldman GH. 2019. Aspergillus fumigatus calcium-responsive transcription factors regulate cell wall architecture promoting stress tolerance, virulence and caspofungin resistance. PLoS Genet 15:e1008551. doi: 10.1371/journal.pgen.1008551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han Z, Tian L, Pecot T, Huang T, Machiraju R, Huang K. 2012. A signal processing approach for enriched region detection in RNA polymerase II ChIP-seq data. BMC Bioinformatics 13(Suppl 2):S2. doi: 10.1186/1471-2105-13-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mokry M, Hatzis P, Schuijers J, Lansu N, Ruzius FP, Clevers H, Cuppen E. 2012. Integrated genome-wide analysis of transcription factor occupancy, RNA polymerase II binding and steady-state RNA levels identify differentially regulated functional gene classes. Nucleic Acids Res 40:148–158. doi: 10.1093/nar/gkr720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vienken K, Fischer R. 2006. The Zn(II)2Cys6 putative transcription factor NosA controls fruiting body formation in Aspergillus nidulans. Mol Microbiol 61:544–554. doi: 10.1111/j.1365-2958.2006.05257.x. [DOI] [PubMed] [Google Scholar]
- 34.Busch S, Schwier EU, Nahlik K, Bayram O, Helmstaedt K, Draht OW, Krappmann O, Valerius O, Lipscomb WN, Braus G. 2007. An eight-subunit COP9 signalosome with an intact JAMM motif is required for fungal fruit body formation. Proc Natl Acad Sci U S A 104:8089–8094. doi: 10.1073/pnas.0702108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alkahyyat F, Ni M, Kim SC, Yu JH. 2015. The WOPR domain protein OsaA orchestrates development in Aspergillus nidulans. PLoS One 10:e0137554. doi: 10.1371/journal.pone.0137554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee MK, Kwon NJ, Lee IS, Jung S, Kim SC, Yu JH. 2016. Negative regulation and developmental competence in Aspergillus. Sci Rep 6:28874. doi: 10.1038/srep28874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bayram O, Sari F, Braus GH, Irniger S. 2009. The protein kinase ImeB is required for light-mediated inhibition of sexual development and for mycotoxin production in Aspergillus nidulans. Mol Microbiol 71:1278–1295. doi: 10.1111/j.1365-2958.2009.06606.x. [DOI] [PubMed] [Google Scholar]
- 38.Wieser J, Adams TH. 1995. flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes Dev 9:491–502. doi: 10.1101/gad.9.4.491. [DOI] [PubMed] [Google Scholar]
- 39.Seo JA, Guan Y, Yu JH. 2003. Suppressor mutations bypass the requirement of fluG for asexual sporulation and sterigmatocystin production in Aspergillus nidulans. Genetics 165:1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wieser J, Lee BN, Fondon J, III, Adams TH. 1994. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr Genet 27:62–69. doi: 10.1007/BF00326580. [DOI] [PubMed] [Google Scholar]
- 41.Sarikaya-Bayram O, Bayram O, Feussner K, Kim JH, Kim HS, Kaever A, Feussner I, Chae KS, Han DM, Han KH, Braus GH. 2014. Membrane-bound methyltransferase complex VapA-VipC-VapB guides epigenetic control of fungal development. Dev Cell 29:406–420. doi: 10.1016/j.devcel.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 42.Balloy V, Chignard M. 2009. The innate immune response to Aspergillus fumigatus. Microbes Infect 11:919–927. doi: 10.1016/j.micinf.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 43.Han DM, Han YJ, Kim JH, Jahng KY, Chung YS, Chung JH, Chae KS. 1994. Isolation and characterization of NSD mutants in Aspergillus nidulans. Korean J Mycol 22:1–7. [Google Scholar]
- 44.Han KH, Han DM. 2010. Genetics and genomics of sexual development of Aspergillus nidulans, p 115–137. In Machida M, Gomi K (ed), Aspergillus molecular biology and genomics. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 45.Han KH, Han KY, Yu JH, Chae K, Jahng KY, Han DM. 2001. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol Microbiol 41:299–309. doi: 10.1046/j.1365-2958.2001.02472.x. [DOI] [PubMed] [Google Scholar]
- 46.Caplan T, Polvi EJ, Xie JL, Buckhalter S, Leach MD, Robbins N, Cowen LE. 2018. Functional genomic screening reveals core modulators of echinocandin stress responses in Candida albicans. Cell Rep 23:2292–2298. doi: 10.1016/j.celrep.2018.04.084. [DOI] [PubMed] [Google Scholar]
- 47.Damveld RA, Franken A, Arentshorst M, Punt PJ, Klis FM, van den Hondel CA, Ram AFJ. 2008. A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 178:873–881. doi: 10.1534/genetics.107.073148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macdonald D, Thomson DD, Johns A, Contreras Valenzuela A, Gilsenan JM, Lord KM, Bowyer P, Denning DW, Read ND, Bromley MJ. 2018. Inducible cell fusion permits use of competitive fitness profiling in the human pathogenic fungus Aspergillus fumigatus. Antimicrob Agents Chemother 63:e01615-18. doi: 10.1128/AAC.01615-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ries LNA, Rocha MC, de Castro PA, Silva-Rocha R, Silva RN, Freitas FZ, de Assis LJ, Bertolini MC, Malavazi I, Goldman GH. 2017. The Aspergillus fumigatus CrzA transcription factor activates chitin synthase gene expression during the caspofungin paradoxical effect. mBio 8:e00705-17. doi: 10.1128/mBio.00705-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braus GH, Krappman S, Eckert SE. 2002. Sexual development in ascomycetes. Fruit body formation of Aspergillus nidulans, p 215–244. In Osiewacz HD (ed), Molecular biology of fungal development. Marcel Dekker, New York, NY. [Google Scholar]
- 51.Sohn KT, Yoon KS. 2002. Ultrastructural study on the cleistothecium development in Aspergillus nidulans. Mycobiology 30:117–127. doi: 10.4489/MYCO.2002.30.3.117. [DOI] [Google Scholar]
- 52.Cary JW, Harris-Coward PY, Ehrlich KC, Mack BM, Kale SP, Larey C, Calvo AM. 2012. NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot Cell 11:1104–1111. doi: 10.1128/EC.00069-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Son H, Seo Y-S, Min K, Park AR, Lee J, Jin J-M, Lin Y, Cao P, Hong S-Y, Kim E-K, Lee S-H, Cho A, Lee S, Kim M-G, Kim Y, Kim J-E, Kim J-C, Choi GJ, Yun S-H, Lim JY, Kim M, Lee Y-H, Choi Y-D, Lee Y-W. 2011. A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog 7:e1002310. doi: 10.1371/journal.ppat.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teichert I, Nowrousian M, Poggeker S, Kuck U. 2014. The filamentous fungus Sordaria macrospora as a genetic model to study fruiting body development. Adv Genet 87:199–244. doi: 10.1016/B978-0-12-800149-3.00004-4. [DOI] [PubMed] [Google Scholar]
- 55.Yu Y, Amich J, Will C, Eagle CE, Dyer PS, Krappmann S. 2017. The novel Aspergillus fumigatus MAT1-2-4 mating-type gene is required for mating and cleistothecia formation. Fungal Genet Biol 108:1–12. doi: 10.1016/j.fgb.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 56.Yu JH. 2010. Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology 38:229–237. doi: 10.4489/MYCO.2010.38.4.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang JY, Chun J, Jun S-C, Han D-M, Chae K-S, Jahng KY. 2013. The MpkB MAP kinase plays a role in autolysis and conidiation of Aspergillus nidulans. Fungal Genet Biol 61:42–49. doi: 10.1016/j.fgb.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 58.van de Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latge JP. 2017. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol 15:661–674. doi: 10.1038/nrmicro.2017.90. [DOI] [PubMed] [Google Scholar]
- 59.Cramer RA, Jr, Perfect BZ, Pinchai N, Park S, Perlin DS, Asfaw YG, Heitman J, Perfect JR, Steinbach WJ. 2008. Calcineurin target CrzA regulates conidial germination, hyphal growth, and pathogenesis of Aspergillus fumigatus. Eukaryot Cell 7:1085–1097. doi: 10.1128/EC.00086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soriani FM, Malavazi I, da Silva Ferreira ME, Savoldi M, Von Zeska Kress MR, Souza Goldman MH, Loss O, Bignell E, Goldman GH. 2008. Functional characterization of the Aspergillus fumigatus CRZ1 homologue, CrzA. Mol Microbiol 67:1274–1291. doi: 10.1111/j.1365-2958.2008.06122.x. [DOI] [PubMed] [Google Scholar]
- 61.Soriani FM, Malavazi I, Savoldi M, Espeso E, Dinamarco TM, Bernardes LA, Ferreira M, Goldman MHS, Goldman GH. 2010. Identification of possible targets of the Aspergillus fumigatus CRZ1 homologue, CrzA. BMC Microbiol 10:12. doi: 10.1186/1471-2180-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Castro PA, Chen C, de Almeida RS, Freitas FZ, Bertolini MC, Morais ER, Brown NA, Ramalho LN, Hagiwara D, Mitchell TK, Goldman GH. 2014. ChIP-seq reveals a role for CrzA in the Aspergillus fumigatus high-osmolarity glycerol response (HOG) signalling pathway. Mol Microbiol 94:655–674. doi: 10.1111/mmi.12785. [DOI] [PubMed] [Google Scholar]
- 63.Thewes S. 2014. Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot Cell 13:694–705. doi: 10.1128/EC.00038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Espeso EA. 2016. The CRaZy calcium cycle. Adv Exp Med Biol 892:169–186. doi: 10.1007/978-3-319-25304-6_7. [DOI] [PubMed] [Google Scholar]
- 65.Brown NA, Goldman GH. 2016. The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance. J Microbiol 54:243–253. doi: 10.1007/s12275-016-5510-4. [DOI] [PubMed] [Google Scholar]
- 66.Rocha MC, Fabri JH, Franco de Godoy K, Alves de Castro P, Hori JI, Ferreira da Cunha A, Arentshorst M, Ram AF, van den Hondel CA, Goldman GH, Malavazi I. 2016. Aspergillus fumigatus MADS-box transcription factor rlmA is required for regulation of the cell wall integrity and virulence. G3 (Bethesda) 6:2983–3002. doi: 10.1534/g3.116.031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Latgé J-P, Chamilos G. 2019. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev 33:e00140-18. doi: 10.1128/CMR.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li S, Bao D, Yuen G, Harris SD, Calvo AM. 2007. basA regulates cell wall organization and asexual/sexual sporulation ratio in Aspergillus nidulans. Genetics 176:243–253. doi: 10.1534/genetics.106.068239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gastebois A, Clavaud C, Aimanianda V, Latge JP. 2009. Aspergillus fumigatus: cell wall polysaccharides, their biosynthesis and organization. Future Microbiol 4:583–595. doi: 10.2217/fmb.09.29. [DOI] [PubMed] [Google Scholar]
- 70.Sales-Campos H, Tonani L, Cardoso CR, Kress MR. 2013. The immune interplay between the host and the pathogen in Aspergillus fumigatus lung infection. Biomed Res Int 2013:693023. doi: 10.1155/2013/693023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kafer E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv Genet 19:33–131. doi: 10.1016/S0065-2660(08)60245-X. [DOI] [PubMed] [Google Scholar]
- 72.Brock M, Jouvion G, Droin-Bergère S, Dussurget O, Nicola MA, Ibrahim-Granet O. 2008. Bioluminescent Aspergillus fumigatus, a new tool for drug efficiency testing and in vivo monitoring of invasive aspergillosis. Appl Environ Microbiol 74:7023–7035. doi: 10.1128/AEM.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geib E, Gressler M, Viediernikova I, Hillmann F, Jacobsen ID, Nietzsche S, Hertweck C, Brock M. 2016. A non-canonical melanin biosynthesis pathway protects Aspergillus terreus conidia from environmental stress. Cell Chem Biol 23:587–597. doi: 10.1016/j.chembiol.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 74.Semighini CP, Marins M, Goldman MH, Goldman GH. 2002. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcription-PCR assay. Appl Environ Microbiol 68:1351–1357. doi: 10.1128/aem.68.3.1351-1357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzuki Y, Murray SL, Wong KH, Davis MA, Hynes MJ. 2012. Reprogramming of carbon metabolism by the transcriptional activators AcuK and AcuM in Aspergillus nidulans. Mol Microbiol 84:942–964. doi: 10.1111/j.1365-2958.2012.08067.x. [DOI] [PubMed] [Google Scholar]
- 76.Wong KH, Jin Y, Moqtaderi Z. 2013. Multiplex Illumina sequencing using DNA barcoding. Curr Protocol Mol Biol Chapter 7:Unit 7.11. doi: 10.1002/0471142727.mb0711s101. [DOI] [PubMed] [Google Scholar]
- 77.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu S. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dallies N, François J, Paquet V. 1998. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast 14:1297–1306. doi:. [DOI] [PubMed] [Google Scholar]
- 82.Masuko T, Minami A, Iwasaki N, Majima T, Nishimura SI, Lee YC. 2005. Carbohydrate analysis by a phenol–sulfuric acid method in microplate format. Anal Biochem 339:69–72. doi: 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 83.Graham LM, Tsoni SV, Willment JA, Williams DL, Taylor PR, Gordon S, Dennehy K, Brown GD. 2006. Soluble Dectin-1 as a tool to detect beta-glucans. J Immunol Methods 314:164–169. doi: 10.1016/j.jim.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 84.Winkelstroter LK, Bom VL, de Castro PA, Ramalho LN, Goldman MH, Brown NA, Rajendran R, Ramage G, Bovier E, Dos Reis TF, Savoldi M, Hagiwara D, Goldman GH. 2015. High osmolarity glycerol response PtcB phosphatase is important for Aspergillus fumigatus virulence. Mol Microbiol 96:42–54. doi: 10.1111/mmi.12919. [DOI] [PubMed] [Google Scholar]
- 85.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cano LE, Singer-Vermes LM, Vaz CAC, Russo M, Calich VLG. 1995. Pulmonary paracoccidioidomycosis in resistant and susceptible mice: relationship among progression of infection, bronchoalveolar cell activation, cellular immune response, and specific isotype patterns. Infect Immun 63:1777–1783. doi: 10.1128/IAI.63.5.1777-1783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guerra ES, Lee CK, Specht CA, Yadav B, Huang H, Akalin A, Huh JR, Mueller C, Levitz SM. 2017. Central role of IL-23 and IL-17 producing eosinophils as immunomodulatory effector cells in acute pulmonary aspergillosis and allergic asthma. PLoS Pathog 13:e1006175. doi: 10.1371/journal.ppat.1006175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marim FM, Silveira TN, Lima DS, Jr, Zamboni DS. 2010. A method for generation of bone marrow-derived macrophages from cryopreserved mouse bone marrow cells. PLoS One 5:e15263. doi: 10.1371/journal.pone.0015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mech F, Thywissen A, Guthke R, Brakhage AA, Figge MT. 2011. Automated image analysis of the host-pathogen interaction between phagocytes and Aspergillus fumigatus. PLoS One 6:e19591. doi: 10.1371/journal.pone.0019591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The transcriptional profiling for ΔnsdC was investigated using ChIP of RNA polymerase II coupled to DNA sequencing. Short reads were deposited in the NCBI, under GEO accession number GSE148557.