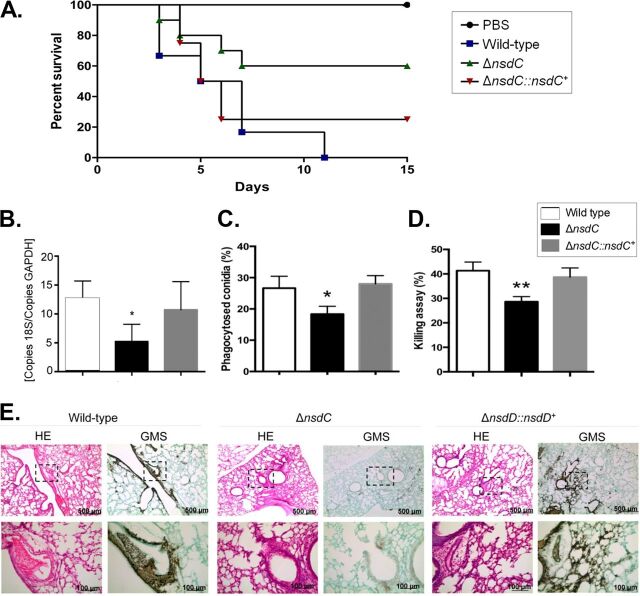
FIG 7.
The A. fumigatus ΔnsdC strain has attenuated virulence in neutropenic mice. (A) Comparative analysis of wild-type, ΔnsdC, and ΔnsdC::nsdC+ strains in a neutropenic murine model of pulmonary aspergillosis. Mice in groups of 10 per strain were inoculated intranasally with a 20-μl suspension of conidia at a dose of 105. PBS, phosphate-buffered saline. The statistical significance of comparative survival values was calculated with the Prism statistical analysis package by using log rank (Mantel-Cox) and Gehan-Brestow-Wilcoxon tests. (B) Fungal burden was determined 48 h postinfection by real-time qPCR based on the 18S rRNA gene of A. fumigatus and an intronic region of the mouse GAPDH gene. Fungal and mouse DNA quantities were obtained from the CT values from an appropriate standard curve. Fungal burden was determined through the ratio between nanograms of fungal DNA and milligrams of mouse DNA. The results are the means (± standard deviations) from five lungs for each treatment. Statistical analysis was performed by using the t test (*, P < 0.05). (C and D) Fungal killing capacity of mouse bone marrow-derived macrophages (BMDMs) assessed as CFU of surviving A. fumigatus strains (wild type, ΔnsdC, or ΔnscD::nscD+ mutant) after 4 h of infection. The percent killing was calculated from CFU remaining compared with control samples without macrophages. The results are the means (± standard deviation) of three independent biological repetitions. Statistical analysis was performed using a one-way ANOVA compared to the wild-type condition (*, P < 0.05; **, P < 0.01). (E) Histopathology of mice infected with A. fumigatus wild-type or ΔnsdC or ΔnscD::nscD+ mutant strain. GMS (Grocott’s methenamine silver) and HE (hematoxylin and eosin) staining of lung sections representative of infections. The boxed area from the top row is magnified in the bottom row. Bars, 100 and 500 μm.