FIG 1.
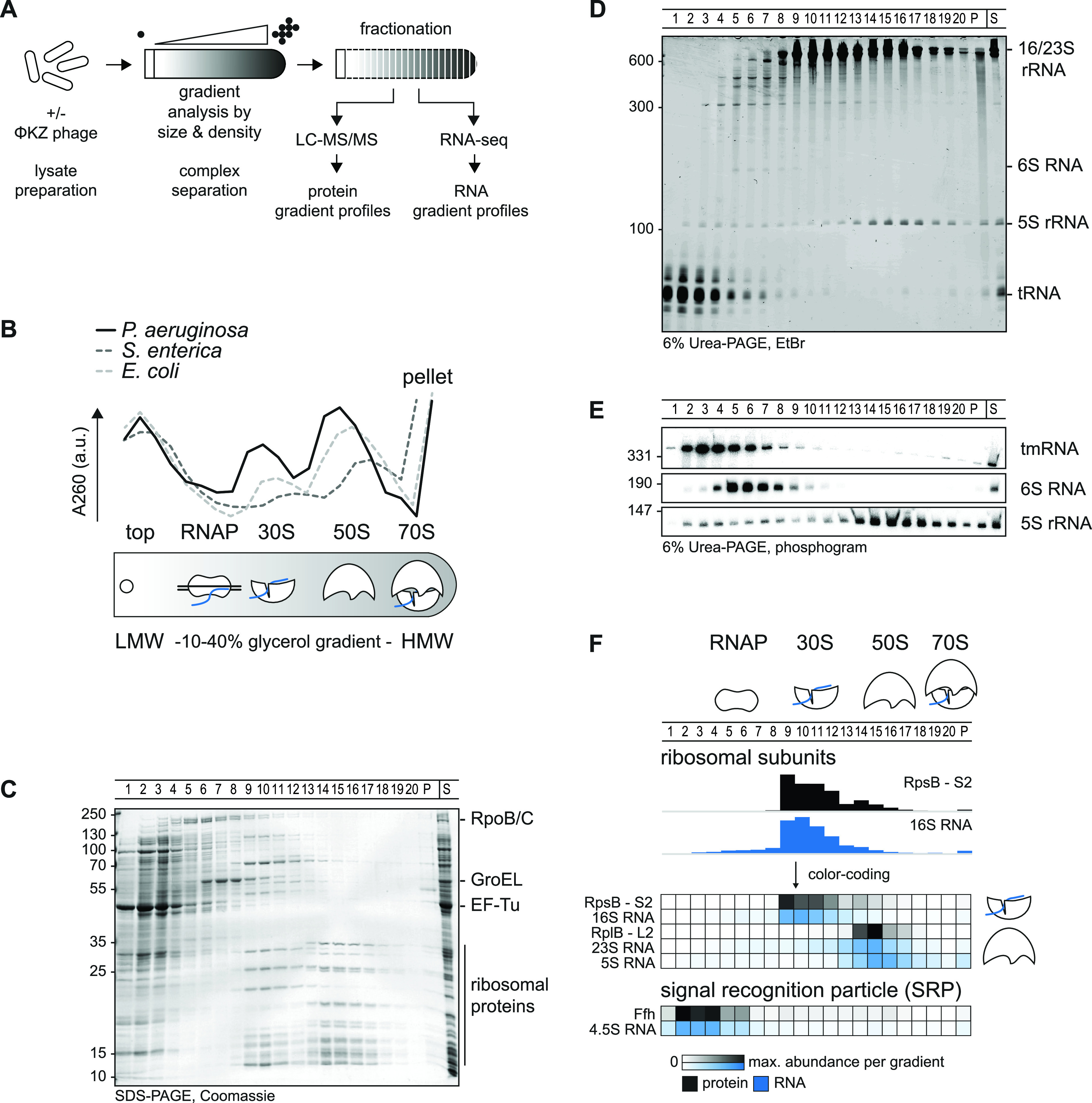
Grad-seq in Pseudomonas. (A) Cellular lysate is analyzed in a gradient and fractionated. High-throughput analysis by mass-spectrometry (LC-MS/MS) and RNA-sequencing reveal sedimentation profiles. (B) Complexes are resolved in a 10% to 40% glycerol gradient by size and density. The 260-nm absorption profile of Pseudomonas lysate indicates LMW fractions at the top, 30S small, 50S large ribosomal subunits at fractions 9 and 15, respectively, and an elevated absorption in the pellet at HMW fractions. The profile is shifted by approximately one fraction compared to those for E. coli and S. enterica. The RNAP sediments between the top and 30S subunit. (C) The apparent proteome resembles the positions in panel B. EF-Tu is a transient translation factor and sedimented in the top; RNAP subunits RpoB/C and GroEL sediment before 30S fractions. (D) tRNAs sedimented in top fractions, ribosomal RNAs resemble well the distribution in panels B and C, and 6S RNA sedimentation correlated with RNAP in panel C. (E) tmRNA sedimented in the pellet and at RNAP fractions; 6S RNA sedimentation correlated with RNAP fractions and 5S rRNA with 50S ribosomal subunit fractions. (F) High-throughput-determined sedimentation profiles matched well with in-gel and previously published results in other species.
