Fig. 1.
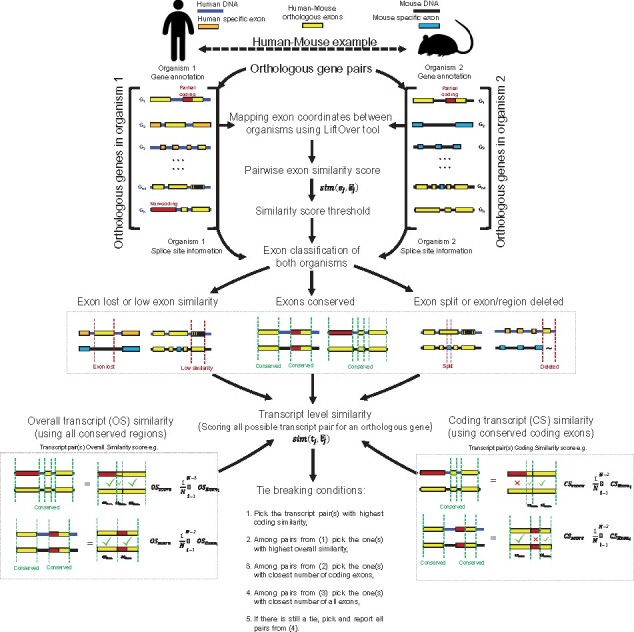
Overview of ExTraMapper. For each orthologous gene pair between the two organisms, ExTraMapper first computes exon-level similarities using gene annotations and the liftover tool that maps regions from one organism to the other. Each exon is then categorized according to its conservation status using the computed similarity scores and splice site information from acceptor and donor sites. The exon-level similarity scores are used to compute transcript-level similarities through an exon pairing algorithm. The transcript-level mappings for each gene are then determined using a greedy method that applies multiple tie break conditions to ensure mostly one-to-one mappings
