Fig. 1.
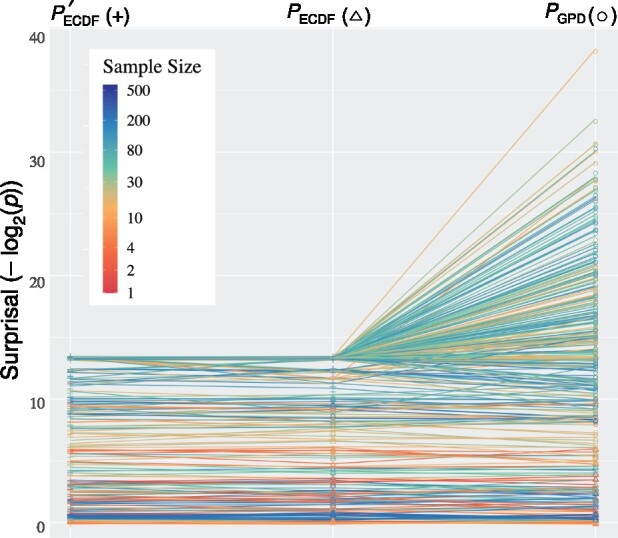
Slopegraph of three different P-value estimate algorithms for KLD CIF divergences of L.enriettii clade tRNA genes (n = 160 genes) against human tRNA genes (n = 431 genes). is the naive Monte Carlo method with 10 000 permutations per feature, using pseudo-counts when there are fewer than S exceedances (by default, 10). terminates after S exceedances () or a maximum of 10 000 permutations, using pseudo-counts unless there are S exceedances. uses algorithm Approximate with T = 500 target permutations and a maximum of 10 000 permutations. Colors show the harmonic mean of the number of sequences containing a given CIF in the two taxa. Points but not lines are jittered against overplotting. See online for color version.
