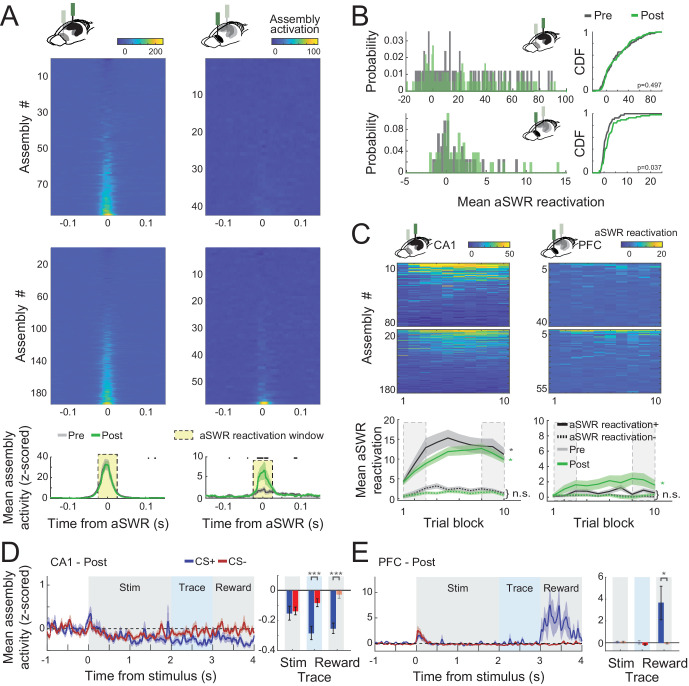
Figure 5. CA1 and prefrontal cortex (PFC) cell assemblies show different awake Sharp-Wave Ripple (aSWR) reactivation dynamics.
(A) Average (z-scored) assembly activation triggered by aSWR occurring in the intertrial intervals for CA1 and PFC, pre- and postlearning sessions (top). Mean aSWR-triggered activation over all the assemblies for pre- and postsessions for each area. Shaded areas represent the standard error of the mean (SEM). Black dots represent windows in which pre- and postassembly activity were statistically different (Wilcoxon rank sum test; p < 0.05). Notice the higher aSWR-triggered activation of assemblies in PFC in postsessions. (B) Histogram (left) and cumulative distribution function (CDF; right) of the mean assembly activity on the reactivation window denoted in A. p values refer to a two-sample Kolmogorov–Smirnov test between pre- and postdistributions. (C) Average aSWR reactivation of each assembly per session (top). Sessions were divided into 10 blocks of equal trial length. Mean aSWR reactivation of all positively (reactivation+) and negatively (reactivation−) reactivated assemblies. Asterisks refer to Wilcoxon signed-rank test performed between the first and last three trial blocks (dashed rectangles) of each area/learning condition (n.s.: nonsignificant; *p < 0.05 and shaded areas represent SEM). Note the evident increase in CA1 aSWR assembly reactivation across the session in both pre- and postsessions for positively modulated assemblies (reactivation+). (D) Mean (z-scored) assembly activity triggered by the stimulus onset for the 25 % most strongly aSWR-reactivated assemblies in CA1 (left). Average of the traces over each trial period is shown for CS+ and CS− (right). Notice the initial decrease of assembly activity in CA1 during the stimulus and the posterior separation between CS+ and CS−. (E) The same as in D, but for PFC assemblies. Note the difference between CS+ and CS− assembly activity during the reward period. Asterisks refer to a Wilcoxon signed-rank test comparing CS+ and CS− (*p < 0.05; ***p < 0.001). Error bars refer to SEM and darker bars denote mean assembly activity significantly different from 0 (p < 0.05; t-test).