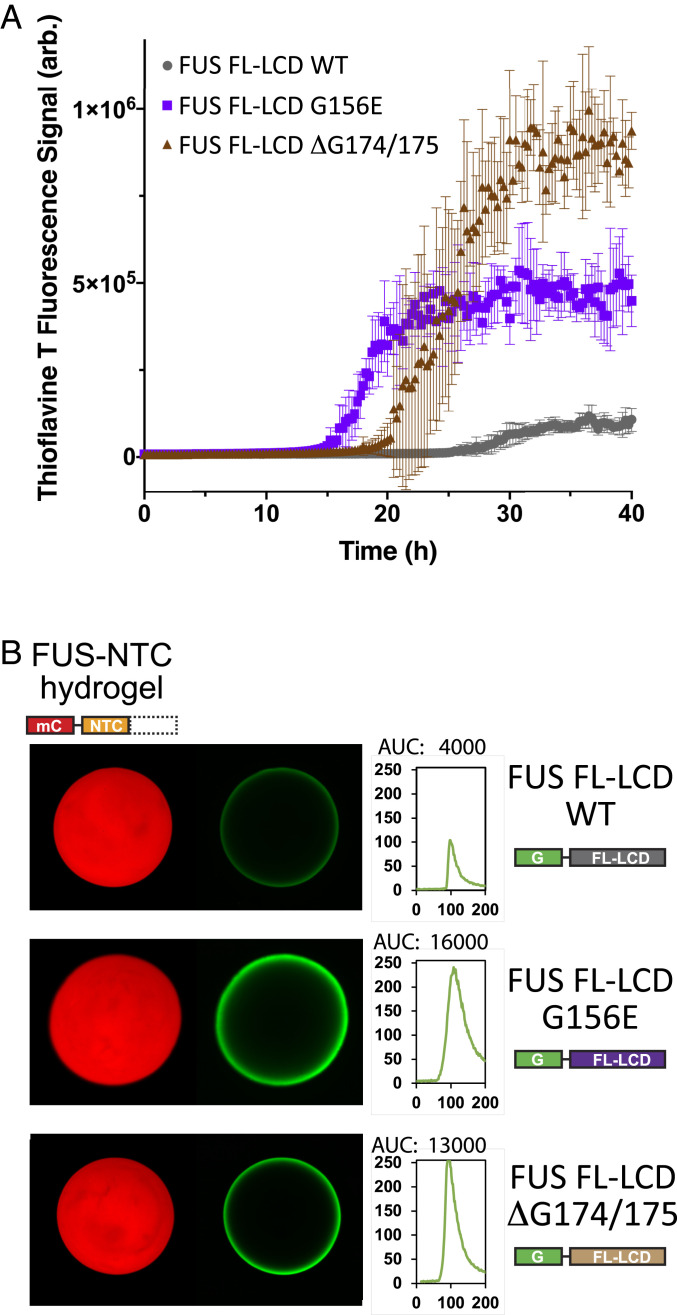
Fig. 7.
Measurements of polymerization and hydrogel binding of ALS-causing mutations as assayed within the intact low-complexity domain of FUS. (A) Full-length derivatives of the FUS low-complexity domain bearing the native amino acid sequence (WT) or that carrying either of two ALS-disposing variants, were expressed as 6His-tagged proteins in bacteria, purified, and incubated under conditions of neutral pH and physiological monovalent salt in the presence of thioflavin-T. The y axis presents thioflavin-T fluorescence; the x axis presents time of incubation. (B) The same three segments of the FUS low-complexity domain were expressed as GFP-tagged proteins in bacteria, purified, and incubated under conditions of neutral pH and physiological monovalent salt with hydrogels formed from mCherry linked to the isolated N-terminal cross-β core of the FUS low-complexity domain. Scans depicted to the Right of hydrogel images present quantitation of GFP signal intensity as measured at hydrogel perimeters. The G156E and ΔG174/G175 variants of the FUS low-complexity domain (Bottom two rows) displayed between three- and fourfold greater binding intensities than that observed for the native FUS protein (Top row).