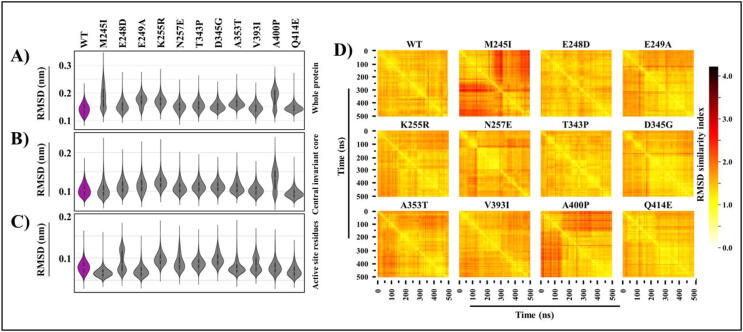
Fig. 2.
Cα backbone stability analysis. A) RMSD violin distribution plots of the A) whole protein, B) central invariant core, and C) binding active site residues of WT and mutant FP-2 proteins. Shown in purple are the WT ensemble plots. D) All vs all Cα RMSD of FP-2 catalytic systems. The × and y-axes denote time (ns). Color scale shows the degree of conformational variability between frames (most similar = white, distinct = dark). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)