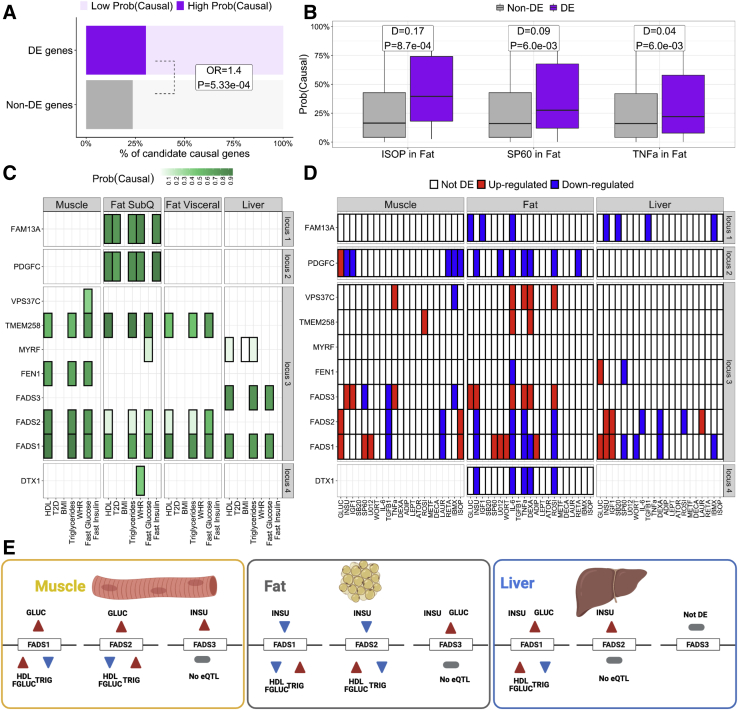
Figure 4.
Environmental perturbations can help inform the functionality of causal genes underlying cardiometabolic trait loci
(A) Percent of causal genes (High Prob(Causal)) underlying cardiometabolic traits loci that are DE (purple) or not DE (gray) in at least one perturbation and cell line. OR/P: odds ratio and Fisher’s exact test p value for the enrichment of DE genes among causal genes compared to non-DE genes.
(B) Perturbation-cell-line combinations with a significant (FDR < 10%) difference in median (D) CLPPmod between DE and non-DE genes, according to the two-samples Wilcoxon rank-sum test.
(C and D) Examples of loci for which intersecting the effects of perturbations (C) with the colocalization results (D) helps inform the functionality of candidate causal genes. The color indicates CLPPmod (C) or DE direction (D). White boxes with crosses indicate that the gene was not tested for colocalization or DE.
(E) Effect of glucose and insulin in the expression of the three FADS genes and the effect of the expression of these genes on HDL, fasting glucose (FGLUC), and triglycerides (TRIG), the three traits for which FADS genes colocalize. The color of the triangles indicates either the effect of the perturbation on the gene (red, upregulation; blue, downregulation) and the effect that up-/downregulation of the gene has on the phenotype (red, increased phenotype, blue, decreased phenotype). DE, differential expression; CLPPmod, LD-adjusted colocalization posterior probability; FDR, false discovery rate.