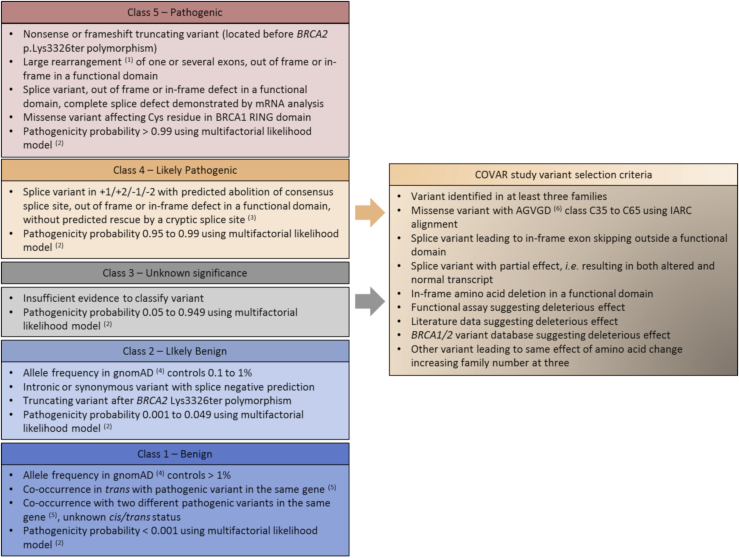
Figure 1.
Variant classification criteria in the framework of the 5 classes of likelihood of pathogenicity and COVAR study variant selection criteria
The variant classification or selection for COVAR study relies on the presence of at least one of the listed criteria.
(1) For large duplications, tandem status disrupting the gene must be demonstrated by mRNA analysis or breakpoint sequencing, otherwise this variant should be considered to be a VUS.
(2) Multifactorial likelihood model proposed by Goldgar et al.17
(3) This classification criterion has been used since January 2019. Before 2019, these splice variants were classified in class 5 and were confirmed by mRNA analysis.
(4) Total gnomAD cohort or subpopulations except for Finnish, Ashkenazi Jewish, and “Other” groups due to the possibility of a founder effect and/or the small number of subjects. gnomAD started to be used as the control database for classification criteria in July 2017. Between December 2014 and June 2017, allele frequency was determined from the ExAC database, with the same thresholds and population groups for classification. Between 2012 and November 2014, allele frequency was determined from the 1000 Genomes Project and Exome Sequencing Project databases, with only one threshold, allele frequency > 1% for classification in class 1 variant, and only the total cohort of both projects.
(5) With no clinical or biological signs of Fanconi anemia.
(6) Align-GVGD prediction algorithm classifies variants from C0 to C65 classes; C0 corresponds to the least likely functional variants and C65 corresponds to the most likely functional variants.31 The last species of IARC alignment, purple sea urchin, is removed for Align-GVGD prediction when it is the only species with an amino acid change; in this case, the inclusion criterion corresponds to Align-GVGD class C35 to C65 AND deleterious prediction by SIFT algorithm.32
IARC, International Agency for Research on Cancer.